What We’re Reading: November 3rd
Review: Plant hormone transporters: what we know and what we would like to know
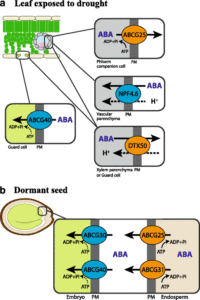 Hormones are signaling molecules, and in most (but not all) cases part of their function is to convey information from one cell or tissue to another, sometimes from cell-to-cell and sometimes through vascular tissues. Park et al. review our current state of understanding of transporters for diverse plant hormones, starting with auxin and extending through ABA, cytokinins, gibberellins, strigolactones, jasmonates, salicylic acid, and ethylene (or more accurately, its precursor ACC), with brassinosteroids, FT and small peptide hormones mentioned briefly. Unresolved questions including the regulation of the transporters are discussed. This article provides a nice overview of the similarities and differences between hormone transport strategies. (Summary by Mary Williams) BMC Biol. 10.1186/s12915-017-0443-x
Hormones are signaling molecules, and in most (but not all) cases part of their function is to convey information from one cell or tissue to another, sometimes from cell-to-cell and sometimes through vascular tissues. Park et al. review our current state of understanding of transporters for diverse plant hormones, starting with auxin and extending through ABA, cytokinins, gibberellins, strigolactones, jasmonates, salicylic acid, and ethylene (or more accurately, its precursor ACC), with brassinosteroids, FT and small peptide hormones mentioned briefly. Unresolved questions including the regulation of the transporters are discussed. This article provides a nice overview of the similarities and differences between hormone transport strategies. (Summary by Mary Williams) BMC Biol. 10.1186/s12915-017-0443-x
Review: Balancing immunity and yield in crop plants
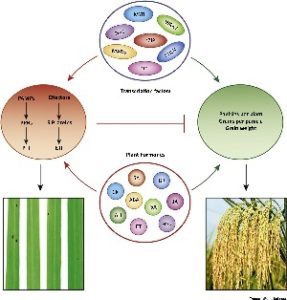 During pathogen invasion, plant resources are diverted from further growth to activation of plant immune response. This review focuses on the recent advances in understanding the complex relationship between immunity regulation and yield production in the model monocot crop rice (Oryza sativa). Many defense-response hormone signaling pathways are discussed, including salicylic acid (SA), brassinosteroid, jasmonates, ethylene, gibberellic acid (GA), abscisic acid, and auxin. In rice, SA signaling interferes with the auxin pathway, leading to enhanced resistance to a bacterial blight pathogen at the expense of plant growth. JA balances activation of pathogen resistance versus rice spikelet development, as seen during plant shade avoidance response. Both SA and JA pathways can be integrated by Slender Rice1 (SLR1) but SLR1 is degraded by upregulation of the growth hormone GA. Just as these examples show the complexity and competition between plant defense hormones and plant growth hormones, transcription factors (TF) also act to regulate plant growth versus plant immunity. By optimizing TF gene expression, an optimal balance between these competing factors can be achieved and may serve as a target for future breeding strategies. On the other hand, less is known about the regulation of resistance genes and the interaction between their subsequent protein products in rice, but work in Arabidopsis shows promise of alleviating the fitness costs associated with their expression. (Summary by Alecia Biel) Trends Plant Sci. 10.1016/j.tplant.2017.09.010.
During pathogen invasion, plant resources are diverted from further growth to activation of plant immune response. This review focuses on the recent advances in understanding the complex relationship between immunity regulation and yield production in the model monocot crop rice (Oryza sativa). Many defense-response hormone signaling pathways are discussed, including salicylic acid (SA), brassinosteroid, jasmonates, ethylene, gibberellic acid (GA), abscisic acid, and auxin. In rice, SA signaling interferes with the auxin pathway, leading to enhanced resistance to a bacterial blight pathogen at the expense of plant growth. JA balances activation of pathogen resistance versus rice spikelet development, as seen during plant shade avoidance response. Both SA and JA pathways can be integrated by Slender Rice1 (SLR1) but SLR1 is degraded by upregulation of the growth hormone GA. Just as these examples show the complexity and competition between plant defense hormones and plant growth hormones, transcription factors (TF) also act to regulate plant growth versus plant immunity. By optimizing TF gene expression, an optimal balance between these competing factors can be achieved and may serve as a target for future breeding strategies. On the other hand, less is known about the regulation of resistance genes and the interaction between their subsequent protein products in rice, but work in Arabidopsis shows promise of alleviating the fitness costs associated with their expression. (Summary by Alecia Biel) Trends Plant Sci. 10.1016/j.tplant.2017.09.010.
Review: The genetics of drought tolerance in conifers
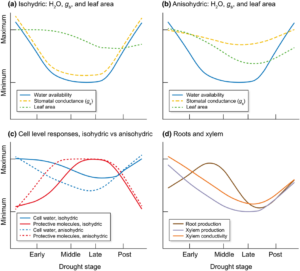 Changing climates mean changing rainfall patterns, which can have serious consequences for long-lived plants such as conifers. Moran et al. provide a thoughtful and readable overview of the strategies that enable some conifer species to survive drought. They start by discussing the different definitions of “drought tolerance” and what they mean for different types of trees (specifically, isohydric, those that close their stomata to maintain a relatively constant hydration state, and anioshydric, those that experience a much greater change in hydration state). What’s particularly nice about this review is that it connects physiological studies of drought tolerance to genetic studies, which is now possible due to the increasing amount of data from conifers. A key recommendation is that researchers focus more on drought survival or growth after drought rather than water-use efficiency (including δ13C) as a measure of drought tolerance. Encouragingly, they conclude, “early attempts at predicting phenotypes from genotypes suggest that genetic tools may be able to aid managers to select appropriate planting stock in the near future, at least for the better studied species.” (Summary by Mary Williams) New Phytol. 10.1111/nph.14774
Changing climates mean changing rainfall patterns, which can have serious consequences for long-lived plants such as conifers. Moran et al. provide a thoughtful and readable overview of the strategies that enable some conifer species to survive drought. They start by discussing the different definitions of “drought tolerance” and what they mean for different types of trees (specifically, isohydric, those that close their stomata to maintain a relatively constant hydration state, and anioshydric, those that experience a much greater change in hydration state). What’s particularly nice about this review is that it connects physiological studies of drought tolerance to genetic studies, which is now possible due to the increasing amount of data from conifers. A key recommendation is that researchers focus more on drought survival or growth after drought rather than water-use efficiency (including δ13C) as a measure of drought tolerance. Encouragingly, they conclude, “early attempts at predicting phenotypes from genotypes suggest that genetic tools may be able to aid managers to select appropriate planting stock in the near future, at least for the better studied species.” (Summary by Mary Williams) New Phytol. 10.1111/nph.14774
Review: New molecular mechanisms to reduce arsenic in crops ($)
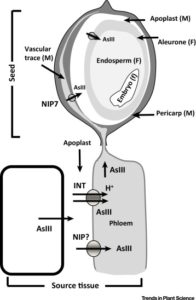 “Over 200 million humans are at risk of arsenic poisoning,” due to arsenic in groundwater and its uptake into crops. Our understanding of the transporters through which arsenic enters the plant, moves through the plant, and enters the seed has increased substantially in recent years, opening the door to diverse strategies for reducing arsenic accumulation in food, as reviewed by Lindsay and Maathuis. (Summary by Mary Williams) Trends Plant Sci. 10.1016/j.tplants.2017.09.015
“Over 200 million humans are at risk of arsenic poisoning,” due to arsenic in groundwater and its uptake into crops. Our understanding of the transporters through which arsenic enters the plant, moves through the plant, and enters the seed has increased substantially in recent years, opening the door to diverse strategies for reducing arsenic accumulation in food, as reviewed by Lindsay and Maathuis. (Summary by Mary Williams) Trends Plant Sci. 10.1016/j.tplants.2017.09.015
Base-editing in RNA and DNA
The ability to engineer precise changes in nucleic acid sequences has advanced rapidly over the last few years. Since the development of genome editing technologies such as CRISPR-Cas9, a modified version known as base editing has sought to reliably convert individual nucleotides. All known base editing techniques harness cytidine deaminases fused to a catalytically inactive Cas9 to permit C•G to T•A conversions. Gaudelli et al. (Nature 10.1038/nature24644) created a modified version of an E. coli adenosine deaminase through several rounds of bacterial evolution that was able to mediate specific A•T to G•C conversions at high efficiency while minimizing off-target effects. As DNA base editors can target both the sense and anti-sense strands, it is now possible to mediate all four base transitions in genomic DNA.
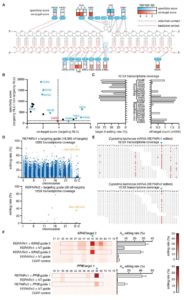 Cox et al. (Science 10.1126/science.aaq0180) used a similar approach to repurpose the RNA-binding Cas13 protein to perform base editing within RNA transcripts. They developed a version of Cas13 without nuclease activity and fused it to an adenosine deaminase acting on RNA (ADAR) family protein. This fusion was capable of directing adenosine to inosine (read as guanosine) replacements within a target site. The ADAR protein was subject to bacterial evolution to minimize non-specific edits around the target site. While currently lacking the range of DNA base editors, the non-permanency of RNA editing is advantageous for some applications.
Cox et al. (Science 10.1126/science.aaq0180) used a similar approach to repurpose the RNA-binding Cas13 protein to perform base editing within RNA transcripts. They developed a version of Cas13 without nuclease activity and fused it to an adenosine deaminase acting on RNA (ADAR) family protein. This fusion was capable of directing adenosine to inosine (read as guanosine) replacements within a target site. The ADAR protein was subject to bacterial evolution to minimize non-specific edits around the target site. While currently lacking the range of DNA base editors, the non-permanency of RNA editing is advantageous for some applications.
These tools are likely to help shape agronomic research in the near future. (Summaries by Mike Page).
The AraGWAS Catalog: a curated and standardized Arabidopsis thaliana GWAS catalog
 AraGWAS (aragwas.1001genomes.org) is a new useful tool developed within the 1001 Genome project that fully integrates and complements GWAP, EasyGWAS, AraPheno and AraGeno. With the availability of the 1001 genomes of Arabidopsis, the number of association studies is dramatically increasing but a way to browse, compare and understand possible connections among them was missing. In their paper, Togninalli and colleagues describe a user-friendly website that catalogues available association studies performed in Arabidopsis and allows browsing through public phenotypes, studies and single nucleotide polymorphism associations. As the GWAS contained in the website have been re-computed with standard methodologies, we can now easily visualize and compare results and statistics of multiple phenotypes, with new possibilities of research and analysis. (Summary by Marco Giovanetti) Nucl. Acid. Res. 10.1093/nar/gkx954
AraGWAS (aragwas.1001genomes.org) is a new useful tool developed within the 1001 Genome project that fully integrates and complements GWAP, EasyGWAS, AraPheno and AraGeno. With the availability of the 1001 genomes of Arabidopsis, the number of association studies is dramatically increasing but a way to browse, compare and understand possible connections among them was missing. In their paper, Togninalli and colleagues describe a user-friendly website that catalogues available association studies performed in Arabidopsis and allows browsing through public phenotypes, studies and single nucleotide polymorphism associations. As the GWAS contained in the website have been re-computed with standard methodologies, we can now easily visualize and compare results and statistics of multiple phenotypes, with new possibilities of research and analysis. (Summary by Marco Giovanetti) Nucl. Acid. Res. 10.1093/nar/gkx954
Embryonic epigenetic reprogramming by a pioneer transcription factor in plants ($)
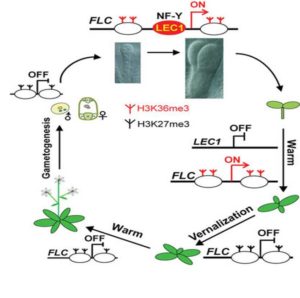 Pioneer transcription factors are a special type of transcription factor that are able to access their target sequences in condensed chromatin. They are often associated with changes in cell fate and developmental switching. Tao et al. showed that the seed-specific transcription factor LEAFY COTYLEDON1 (LEC1) acts as a pioneer transcription factor. In Arabidopsis, prolonged cold leads to the epigenetic silencing of the flowering suppressor FLC, thus enabling the plant to flower, but FLC is reset to an active state during embryogenesis. This new work shows that LEC1 is necessary for FLC resetting during embryo formation and is necessary for the increase in activating histone marks at the FLC locus. (Summary by Mary Williams) Nature 10.1038/nature24300
Pioneer transcription factors are a special type of transcription factor that are able to access their target sequences in condensed chromatin. They are often associated with changes in cell fate and developmental switching. Tao et al. showed that the seed-specific transcription factor LEAFY COTYLEDON1 (LEC1) acts as a pioneer transcription factor. In Arabidopsis, prolonged cold leads to the epigenetic silencing of the flowering suppressor FLC, thus enabling the plant to flower, but FLC is reset to an active state during embryogenesis. This new work shows that LEC1 is necessary for FLC resetting during embryo formation and is necessary for the increase in activating histone marks at the FLC locus. (Summary by Mary Williams) Nature 10.1038/nature24300
Cytokinin-induced cell cycle regulates MET1 activity during shoot regeneration ($)
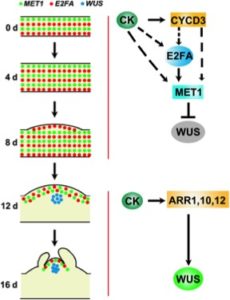 The capacity of plants to regenerate new shoots from differentiated tissue – a process called de novo shoot regeneration – confers plasticity to plant development and has also important agricultural applications. Previous studies revealed that DNA METHYLTRANSFERASE1 (MET1) inhibits shoot regeneration through the repression of the shoot initiation-regulator WUSCHEL (WUS). However, upstream regulators of MET1 expression are unknown. Using a combination of genetics, confocal microscopy and molecular biology in Arabidopsis, Liu et al. reveal that MET1 expression is spatially dynamic during shoot regeneration and fine-tunes WUS expression through methylation of its promoter region. Moreover, MET1 expression is enhanced by the cell cycle regulator E2F as well as by the CYCD3 cyclin in a cytokinin-dependent fashion. Altogether, the authors suggest a model where WUS expression is tightly controlled by the local balance between the repressing CK-CYCD3-E2F-MET1 module and the activating cytokinin response factors. These findings reveal a direct epigenetic control of plant regeneration regulated by external signals including phytohormones. (Summary by Matthias Beniot) New Phytol. 10.1111/nph.14814/full
The capacity of plants to regenerate new shoots from differentiated tissue – a process called de novo shoot regeneration – confers plasticity to plant development and has also important agricultural applications. Previous studies revealed that DNA METHYLTRANSFERASE1 (MET1) inhibits shoot regeneration through the repression of the shoot initiation-regulator WUSCHEL (WUS). However, upstream regulators of MET1 expression are unknown. Using a combination of genetics, confocal microscopy and molecular biology in Arabidopsis, Liu et al. reveal that MET1 expression is spatially dynamic during shoot regeneration and fine-tunes WUS expression through methylation of its promoter region. Moreover, MET1 expression is enhanced by the cell cycle regulator E2F as well as by the CYCD3 cyclin in a cytokinin-dependent fashion. Altogether, the authors suggest a model where WUS expression is tightly controlled by the local balance between the repressing CK-CYCD3-E2F-MET1 module and the activating cytokinin response factors. These findings reveal a direct epigenetic control of plant regeneration regulated by external signals including phytohormones. (Summary by Matthias Beniot) New Phytol. 10.1111/nph.14814/full
Chlorophyll can be reduced in crop canopies with little penalty to photosynthesis
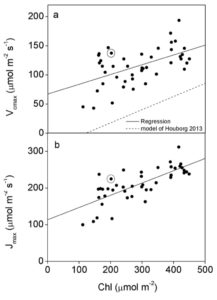 The effect of reducing leaf chlorophyll content on canopy CO2 assimilation (Acan) is somewhat contentious. Walker et al. obtained data from 67 soybean accessions to parameterise a canopy-root-soil model (MLCan) in order to simulate the effect of altering chlorophyll levels on Acan. There was no increase in Acan when chlorophyll content was reduced – while lower canopy layers had elevated assimilation in pale green lines, upper layers performed less well than dark green counterparts. Paler lines also exhibited an increase in lost PPFD through leaf reflectance, while leaf biochemical capacity did not match the redistribution of PPFD throughout the canopy. However, encouragingly there was little detrimental effect on Acan of even very large reductions in chlorophyll, indicating that nitrogen could be repartitioned to the benefit of limiting processes in soybean leaves. These results will be invaluable in formulating a strategy to increase Acan in dense canopy crops such as soybean. (Summary by Mike Page) Plant Physiol.
The effect of reducing leaf chlorophyll content on canopy CO2 assimilation (Acan) is somewhat contentious. Walker et al. obtained data from 67 soybean accessions to parameterise a canopy-root-soil model (MLCan) in order to simulate the effect of altering chlorophyll levels on Acan. There was no increase in Acan when chlorophyll content was reduced – while lower canopy layers had elevated assimilation in pale green lines, upper layers performed less well than dark green counterparts. Paler lines also exhibited an increase in lost PPFD through leaf reflectance, while leaf biochemical capacity did not match the redistribution of PPFD throughout the canopy. However, encouragingly there was little detrimental effect on Acan of even very large reductions in chlorophyll, indicating that nitrogen could be repartitioned to the benefit of limiting processes in soybean leaves. These results will be invaluable in formulating a strategy to increase Acan in dense canopy crops such as soybean. (Summary by Mike Page) Plant Physiol.
ABA-induced reactive oxygen species are modulated by flavonols to control stomata aperture
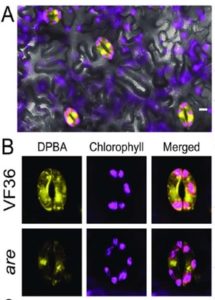 Much of our knowledge concerning ABA-induced stomatal closure comes from genetic models such as Arabidopsis and Vicia faba. Watkins et al. explore the mechanism of ROS production in this abiotic stress pathway in an important agricultural crop: tomatoes (Solanum lycopersicum). Specifically, they are interested in the role of flavonoids in guard cell dynamics of S. lycopersicum. Through a combination of genetic analysis utilizing two mutants displaying elevated or depleted flavonol levels, in addition to hormone treatment assays, it was determined that flavonoids act as antioxidizing agents and flavonoids are upregulated in the presence of ethylene. Flavonols counteracted ABA action by decreasing ROS accumulation. As ROS is required for ABA-induced stomatal closure, enhanced flavonol synthesis inhibited ABA-induced stomatal closure and was also shown to enhance stomatal opening. The inverse relationship was also seen in that less flavonol synthesis results in greater ROS accumulation and enhanced stomatal closure. Agricultural crop production is becoming increasingly important as Earth’s surface temperatures continue to rise and food availability becomes threatened. (Summary by Alecia Biel) Plant Phys. 10.1104/pp.17.01010.
Much of our knowledge concerning ABA-induced stomatal closure comes from genetic models such as Arabidopsis and Vicia faba. Watkins et al. explore the mechanism of ROS production in this abiotic stress pathway in an important agricultural crop: tomatoes (Solanum lycopersicum). Specifically, they are interested in the role of flavonoids in guard cell dynamics of S. lycopersicum. Through a combination of genetic analysis utilizing two mutants displaying elevated or depleted flavonol levels, in addition to hormone treatment assays, it was determined that flavonoids act as antioxidizing agents and flavonoids are upregulated in the presence of ethylene. Flavonols counteracted ABA action by decreasing ROS accumulation. As ROS is required for ABA-induced stomatal closure, enhanced flavonol synthesis inhibited ABA-induced stomatal closure and was also shown to enhance stomatal opening. The inverse relationship was also seen in that less flavonol synthesis results in greater ROS accumulation and enhanced stomatal closure. Agricultural crop production is becoming increasingly important as Earth’s surface temperatures continue to rise and food availability becomes threatened. (Summary by Alecia Biel) Plant Phys. 10.1104/pp.17.01010.
ABA accumulation in dehydrating leaves is associated with decline in cell volume not turgor pressure
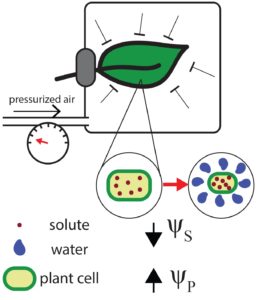
Desiccating leaves show increased ABA levels triggered by low turgor – right? Apparently not! The pressure chamber experiments showing increased ABA levels in desiccating leaves are inconsistent when the entire leaves are enclosed in the chamber. Sack et al. proposes that the turgor pressure is increased during the pressure chamber treatment, rather than decreased and could therefore not be a trigger for ABA accumulation. New equations allow estimation of relative water content, which corresponds to reduction in cell volume and ABA accumulation. Applying the proposed equations to new high-resolution pressure studies will allow identification of dehydration sensors, based on decline in relative water content, occurring before destructive turgor loss. (Summary by Magdalena Julkowska) Plant Physiol.
Natural variation identified genes affecting drought-induced ABA accumulation in Arabidopsis thaliana ($)
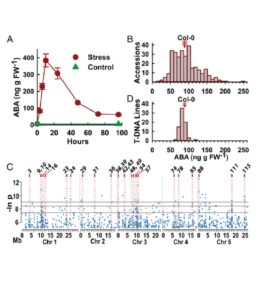
Dissecting the genetic controls of ABA is challenging, granted the complex modulation of ABA turnover and redundancy of the hormone perception machinery. In a recent study, Kalladan and colleagues explored natural variation of low-water-potential induced ABA accumulation. Using heuristic GWAS, followed by a reverse genetic screen, the team identified 116 ABA effector loci. The identified genes often encode signaling protein associated with the plasma membrane, including RLK and ATAucsia-1. The effect of any single T-DNA line studied was remarkably small compared to observed natural variation, suggesting that control of ABA levels is divided among many genes, each with an incremental effect. (Summary by Magdalena Julkowska) Proc. Natl. Acad. Sci. USA 10.1073/pnas.1705884114
Deep learning for multi-task plant phenotyping
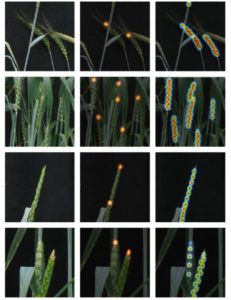 Despite the significant developments in computational biology and modern plant breeding, crop phenotyping poses challenges for automation. Previous machine learning approaches of plant phenotyping have mainly focused on leaf counting in rosette forms or leaf segmentation, and rely on large datasets not particularly useful for monocot crops. Pound et al. have recently described a valuable dataset tool called ACID (The Annotated Crop Image Dataset) that is primarily developed on wheat to localize spikes and spikelets. The dataset is based on a representation of 520 images of wheat plants with a wide range of plant and spike phenotypes. Each plant has been annotated individually to determine the location of spike and individual spikelets. The network performs simultaneous classification of images, with a detailed annotation of spike and spikelets and classification of awned plants. This is an interesting advancement in machine learning phenotyping (http://plantimages.nottingham.ac.uk/) and will be useful for scientists in the improvement of crop phenotyping on all plants with architecture similar to wheat. With the advancement of wheat breeding with varied phenotypic lines, the deep learning machine approach will be necessary for accurate phenotypic quantification with a view to predict yield. (Summary by Amey Redkar) bioRxiv 10.1101/204552
Despite the significant developments in computational biology and modern plant breeding, crop phenotyping poses challenges for automation. Previous machine learning approaches of plant phenotyping have mainly focused on leaf counting in rosette forms or leaf segmentation, and rely on large datasets not particularly useful for monocot crops. Pound et al. have recently described a valuable dataset tool called ACID (The Annotated Crop Image Dataset) that is primarily developed on wheat to localize spikes and spikelets. The dataset is based on a representation of 520 images of wheat plants with a wide range of plant and spike phenotypes. Each plant has been annotated individually to determine the location of spike and individual spikelets. The network performs simultaneous classification of images, with a detailed annotation of spike and spikelets and classification of awned plants. This is an interesting advancement in machine learning phenotyping (http://plantimages.nottingham.ac.uk/) and will be useful for scientists in the improvement of crop phenotyping on all plants with architecture similar to wheat. With the advancement of wheat breeding with varied phenotypic lines, the deep learning machine approach will be necessary for accurate phenotypic quantification with a view to predict yield. (Summary by Amey Redkar) bioRxiv 10.1101/204552
Microbial landscape of the grapevine endosphere in the context of Pierce’s disease
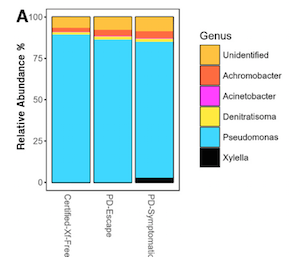 In vascular plants, structure and composition of microbial endosphere associations has not been studied much in the specific context of vascular diseases. This is the case of Pierce’s disease which is caused by Xylella fastidiosa and affects multiple crops and ornamental plants. This bacterium is currently regarded as a major threat in Europe, affecting very important crops such as grapevines and olive trees in Italy, Spain and France. Deyett et al. hypothesized that the differential microbial community inhabiting the grapevine vascular endosphere is a major contributor to resistance in clonal vines, otherwise genetically identical, which are under high diseases pressure in the field. They compared the microbiome residing in clonal grapevines that displayed severely to mildly and asymptomatic disease symptoms by using next-generation sequencing Illumina MiSeq-based platform. Pseudomonas fluorescens and Achromobacter xylosoxidans appeared linked to the asymptomatic phenotype and, therefore, are regarded as putative biological control agents of the disease. (Summary by Isabel Mendoza) Phytobiomes J. 10.1094/PBIOMES-08-17-0033-R
In vascular plants, structure and composition of microbial endosphere associations has not been studied much in the specific context of vascular diseases. This is the case of Pierce’s disease which is caused by Xylella fastidiosa and affects multiple crops and ornamental plants. This bacterium is currently regarded as a major threat in Europe, affecting very important crops such as grapevines and olive trees in Italy, Spain and France. Deyett et al. hypothesized that the differential microbial community inhabiting the grapevine vascular endosphere is a major contributor to resistance in clonal vines, otherwise genetically identical, which are under high diseases pressure in the field. They compared the microbiome residing in clonal grapevines that displayed severely to mildly and asymptomatic disease symptoms by using next-generation sequencing Illumina MiSeq-based platform. Pseudomonas fluorescens and Achromobacter xylosoxidans appeared linked to the asymptomatic phenotype and, therefore, are regarded as putative biological control agents of the disease. (Summary by Isabel Mendoza) Phytobiomes J. 10.1094/PBIOMES-08-17-0033-R
Earth’s very first trees ($)
 Long ago, the dinosaurs roamed amongst majestic forests of ancient tree ferns, cycads and conifers. But longer ago still, prior to the birth of both the kings of the animal and plant kingdoms, majestic forests of gigantic trees were unimaginable, with the landscape covered in small plants lacking leaves, flowers, and roots. As time passed, the first plants awarded the title of tree appeared, although it has long been uncertain exactly how large these species were, and how they could even grow large, as the structures of modern giants (secondary wood laid down concentrically) originated later. A recent study by Xu et al. revealed exceptionally preserved fossils of the earliest trees, Cladoxylopsida, which appeared during the early Mid-Devonian, ~393 million years ago. These trees did not contain the modern means of wood development to grow as large as they did; trunks of up to 70cm in diameter likely capable of supporting long tapering trunks. In contrast to secondary wood growth that supports the large statures of most modern trees, these ancient giants had structures more like that of modern palms, with a network of primary xylem strands embedded within parenchyma, but completely lacking secondary growth. Although arborescent monocots and these early trees likely evolved their comparable features independently, it is striking that ancient species already utilized forms recognizable in modern plants. (Summary by Danielle Roodt Prinsloo) Proc. Natl. Acad. Sci. USA 10.1073/pnas.1708241114.
Long ago, the dinosaurs roamed amongst majestic forests of ancient tree ferns, cycads and conifers. But longer ago still, prior to the birth of both the kings of the animal and plant kingdoms, majestic forests of gigantic trees were unimaginable, with the landscape covered in small plants lacking leaves, flowers, and roots. As time passed, the first plants awarded the title of tree appeared, although it has long been uncertain exactly how large these species were, and how they could even grow large, as the structures of modern giants (secondary wood laid down concentrically) originated later. A recent study by Xu et al. revealed exceptionally preserved fossils of the earliest trees, Cladoxylopsida, which appeared during the early Mid-Devonian, ~393 million years ago. These trees did not contain the modern means of wood development to grow as large as they did; trunks of up to 70cm in diameter likely capable of supporting long tapering trunks. In contrast to secondary wood growth that supports the large statures of most modern trees, these ancient giants had structures more like that of modern palms, with a network of primary xylem strands embedded within parenchyma, but completely lacking secondary growth. Although arborescent monocots and these early trees likely evolved their comparable features independently, it is striking that ancient species already utilized forms recognizable in modern plants. (Summary by Danielle Roodt Prinsloo) Proc. Natl. Acad. Sci. USA 10.1073/pnas.1708241114.