Application of TurboID-mediated proximity labeling for mapping a GSK3 Kinase signaling network in Arabidopsis (bioRxiv)
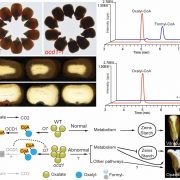
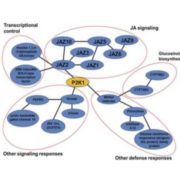
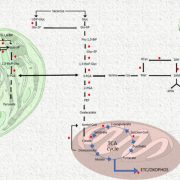
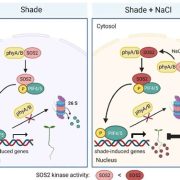
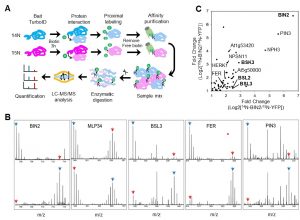 In this preprint Kim et al., have provided an optimized method for determining protein-protein interaction in plants using TurboID-mediated labelling. TurboID is an engineered promiscuous biotin ligase that marks proteins it comes in contact with. Identification of interacting partners of a protein through this approach will aid in deciphering the molecular signaling network. In this approach the protein of interest is tagged with a fluorescent marker by TurboID fused to a protein of interest, driven by a constitutive promoter. This allows all the proteins that comes close to the protein of interest to be biotinylated by TurboID more efficiently so that they can can be detected via pull down assays and mass spectrometry. To evaluate the efficiency of this assay in Arabidopsis the authors have identified the interaction partners of brassinosteroid signaling component, BIN2 (GSK3 like kinase) which will uncover the signaling cascade of this phytohormone. (Summary by Suresh Damodaran) bioRxiv 10.1101/636324
In this preprint Kim et al., have provided an optimized method for determining protein-protein interaction in plants using TurboID-mediated labelling. TurboID is an engineered promiscuous biotin ligase that marks proteins it comes in contact with. Identification of interacting partners of a protein through this approach will aid in deciphering the molecular signaling network. In this approach the protein of interest is tagged with a fluorescent marker by TurboID fused to a protein of interest, driven by a constitutive promoter. This allows all the proteins that comes close to the protein of interest to be biotinylated by TurboID more efficiently so that they can can be detected via pull down assays and mass spectrometry. To evaluate the efficiency of this assay in Arabidopsis the authors have identified the interaction partners of brassinosteroid signaling component, BIN2 (GSK3 like kinase) which will uncover the signaling cascade of this phytohormone. (Summary by Suresh Damodaran) bioRxiv 10.1101/636324