Precise integration of large DNA sequences in plant genomes using PrimeRoot editors
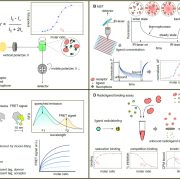
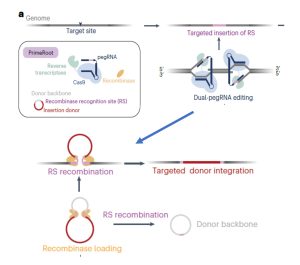 Although CRISPR/Cas9 tools have provided new opportunities for genome editing, using these systems to introduce large pieces of DNA has been challenging. A new genome editing technique, “PrimeRoot” (Prime editing-mediated Recombination Of Opportune Targets), was introduced by Sun et al. and shown to precisely insert large DNA donors/cargos in rice without double stranded break intermediates. The PrimeRoot technique uses plant-optimized guide RNA along with superior site-specific recombinases and an efficient plant prime editor. The authors surveyed the rice genome and found many genomic safe harbor sites, which are regions that can tolerate large insertions without adverse effects and are physically distant from gene coding regions, small RNAs, microRNAs, promoters, enhancers, long non-coding RNAs, etc. The use of optimized prime editing guide RNA (pegRNA) along with an enhanced plant prime editor led to insertion event efficiency of up to 50% in rice. Using this method, the authors were able to integrate a 4.9kb donor cassette comprising PigmR, which confers rice blast disease resistance, driven by the Actin1 promoter into one of the genomic safe harbor sites. The resulting plants showed significantly increased disease resistance, thus demonstrating PrimeRoot’s precision and efficiency for future molecular breeding efforts. (Summary by Indrani Kakati @indranikb) Nature Biotechnol. 10.1038/s41587-023-01769-w
Although CRISPR/Cas9 tools have provided new opportunities for genome editing, using these systems to introduce large pieces of DNA has been challenging. A new genome editing technique, “PrimeRoot” (Prime editing-mediated Recombination Of Opportune Targets), was introduced by Sun et al. and shown to precisely insert large DNA donors/cargos in rice without double stranded break intermediates. The PrimeRoot technique uses plant-optimized guide RNA along with superior site-specific recombinases and an efficient plant prime editor. The authors surveyed the rice genome and found many genomic safe harbor sites, which are regions that can tolerate large insertions without adverse effects and are physically distant from gene coding regions, small RNAs, microRNAs, promoters, enhancers, long non-coding RNAs, etc. The use of optimized prime editing guide RNA (pegRNA) along with an enhanced plant prime editor led to insertion event efficiency of up to 50% in rice. Using this method, the authors were able to integrate a 4.9kb donor cassette comprising PigmR, which confers rice blast disease resistance, driven by the Actin1 promoter into one of the genomic safe harbor sites. The resulting plants showed significantly increased disease resistance, thus demonstrating PrimeRoot’s precision and efficiency for future molecular breeding efforts. (Summary by Indrani Kakati @indranikb) Nature Biotechnol. 10.1038/s41587-023-01769-w