An Improved Recombineering Toolset for Plants
Brumos and Zhao et al. developed an improved set of genetic tools to make precise sequence modifications in large DNA fragments and used them to tag hundreds of plant genes, including those involved in auxin production. Plant Cell https://doi.org/10.1105/tpc.19.00431
By Javier Brumos, Anna Stepanova, and Jose Alonso
Department of Plant and Microbial Biology, Program in Genetics, North Carolina State University, Raleigh, NC, USA
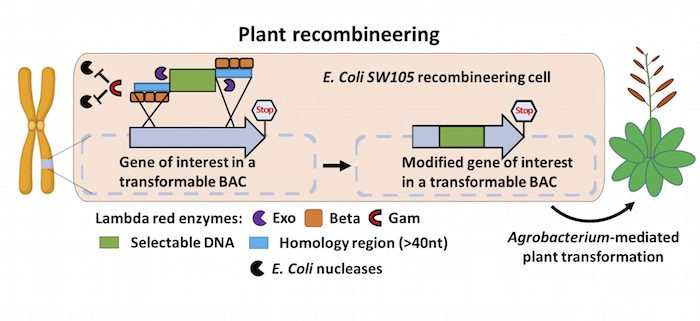
Background: In order to sprout, grow, photosynthesize, fight off pests, flower, or produce fruits, plants turn on different sets of genes in their genomes. To understand how these basic processes are brought about, scientists must first determine what individual plant genes do. To do this, researchers typically modify genes of interest in the laboratory by changing or disabling specific gene functions via mutation, or fuse DNA to a reporter tag such as GFP. However, manipulating large genes in a test tube is often technically difficult. A technology called recombineering makes working with large (e.g., 100 kilobases) DNA fragments easier, but this method has not been widely adopted in plants because it requires specialized genetic tools.
Question: We aimed to develop genetic tools and laboratory protocols so that any plant biology researcher with basic molecular biology equipment and skills could take advantage of recombineering technology. Such tools and procedures must be customizable, scalable, and freely available to all researchers.
Findings: We developed a toolset that makes recombineering faster and easier. We streamlined the process of identifying the bacterial strain carrying the large DNA fragment with the gene of interest and made the recombineering protocols scalable and applicable to a wide set of plant species. Such flexibility makes any type of precise gene editing possible, from changing a single letter in the DNA code to introducing or removing specific sequences. We employed the recombineering pipeline to generate over 250 whole-gene constructs, characterized transgenic lines for over 50 genes, and made the seeds available through the Arabidopsis Biological Stock Center. We built and characterized a complete set of transgenic lines for genes involved in producing the plant hormone auxin and made all of the tools and procedures publicly available.
Next steps: Our transgenic reporter lines could be used to analyze the auxin biosynthesis pathway in any tissue, developmental stage, genetic background, or environmental or experimental condition. Our optimized recombineering protocols could be used in any transformable plant species. Finally, these tools could be utilized in ultra-high-throughput procedures to extend these types of analyses to every gene in a plant genome.
- Brumos, C. Zhao, Y. Gong, D. Soriano, A.P. Patel, M.A. Perez-Amador, A.N. Stepanova, J.M. Alonso (2019). An Improved Recombineering Toolset for Plants. Plant Cell. https://doi.org/10.1105/tpc.19.00431.
Key words: Arabidopsis, recombineering, gene-expression, auxin