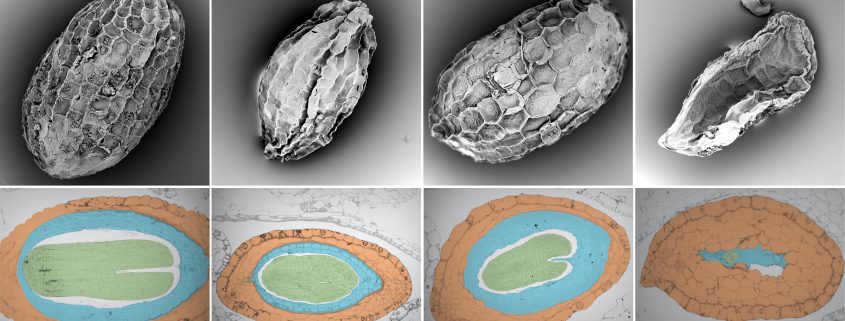
We’re just not compatible…. Plant species with different parenting strategies can’t quite coordinate in rearing hybrid babies
By Taliesin Kinser (University of Florida) and Joshua Puzey (College of William and Mary)
Background: Flowering plants are an evolutionary success story for many reasons, particularly for their unique sex habits. Not only do flowers make long-distance mating possible, seeds also include a portable nutrient source: the endosperm, formed from the other fertilization event and whose sole purpose is to support embryo growth. Uniquely, the endosperm has an extra copy of mom’s genome, and the parental genomes split and coordinate tasks vital for its growth (genomic imprinting). But if parents pass on too many genome copies, or if they are different species entirely, endosperm growth fails, and the embryo perishes. Such failure may involve divergent imprinting roles and methylation marks on DNA – both affecting genome behavior and interactions. Unraveling the process of endosperm failure in hybrid seeds is crucial to understanding endosperm function and how it contributes to flowering plant success and diversity.
Question: We crossed two monkeyflower species, one itself being a hybrid containing two separate genomes (a tetraploid), to test the following: How does endosperm growth contribute to complications in these hybrid seeds? Does the endosperm differ in imprinting roles and DNA methylation between species, and how might these control the parent’s ability to coordinate endosperm growth?
Findings: Hybrid endosperm grows cramped from the start or never takes off, resulting in either shriveled or aborted seeds, respectively, depending on crossing direction. The two species themselves have very different imprinting outcomes and DNA methylation marks in their endosperms. In fact, even though the tetraploid’s two genomes are as genetically distinct as they are to the diploid’s, their imprinting and DNA methylation are much more similar. In hybrid seeds, the tetraploid’s genomes jointly dominate over the diploid genome, likely owing to their differences, overshadowing imprinting roles. Intriguingly, failure of important DNA methylation machinery accompanies hybrid seed abortion, suggesting that methylation and imprinting differences between parents may interrupt their proper coordination, disrupting endosperm, and ultimately seed, development.
Next Steps: We plan to determine the specific functional roles of the identified imprinted genes and examine other genome features in the endosperm of both species and their three genomes within hybrid endosperm to uncover the mechanisms involved.
Reference.
Taliesin J. Kinser, Ronald D. Smith, Amelia H. Lawrence, Arielle M. Cooley, Mario Vallejo-Marín, Gregory D. Conradi Smith, and Joshua R. Puzey (2021). Endosperm-based incompatibilities in hybrid monkeyflowers. Plant Cell. https://doi.org/10.1093/plcell/koab117