Sugars Inform the Circadian Clock How to Shape Rice Shoots via the Strigolactone Pathway
Circadian clocks act as universal timekeepers to harmonize internal processes with external day-night rhythms. Genetic feedback loops gear these inner clocks by approximating time in response to dawn-dusk cycles. In plants, development, growth, hormone action, metabolism and other downstream events are crucially synchronized with the clock (Greenham and McClung, 2015). How the clock shapes plant architecture and yield remain key questions. Here, in an illuminating report, Wang and coworkers (2020) uncover genetic mechanisms in rice by which the circadian clock integrates photosynthetic sugars to regulate the phytohormone strigolactone (SL) pathway, axillary bud potential (i.e., tiller outgrowth), panicle architecture and yield.
 The rice (Oryza sativa) clock is composed of the hub transcription factor CIRCADIAN CLOCK ASSOCIATED1 (OsCCA1) and its antagonist factor PSUEDORESPONSE REGULATOR1 (OsPRR1) that are homologous to Arabidopsis thaliana clock regulators (Murakami et al. 2007). To explore whether tinkering with the clock affects shoot architecture and yield traits, the authors leveraged a series of transgenic experiments (overexpression, gene silencing and CRISPR/Cas9-gene editing) that tested OsCCA1 and OsPRR1 function. Interesting, tiller-bud outgrowth increased or decreased when OsCCA1 was either down- or up-regulated, respectively; altering OsPRR1 levels produced opposite effects. SL is a known inhibitor of rice tillering (Wang et al. 2018). To connect tiller outgrowth with OsCCA1 function, the authors mined published chromatin immunoprecipitation sequencing data for Arabidopsis CCA1 and panned for genes in the SL pathway. Among the putative AtCCA1-SL pathway targets were rice homologs of TEOSINTE BRANCHED1 (OsTB1), DWARF10 (D10), D14 and IDEAL PLANT ARCHITECTURE1 (IPA1), all of which house CCA1-binding sites in their respective promoters.
The rice (Oryza sativa) clock is composed of the hub transcription factor CIRCADIAN CLOCK ASSOCIATED1 (OsCCA1) and its antagonist factor PSUEDORESPONSE REGULATOR1 (OsPRR1) that are homologous to Arabidopsis thaliana clock regulators (Murakami et al. 2007). To explore whether tinkering with the clock affects shoot architecture and yield traits, the authors leveraged a series of transgenic experiments (overexpression, gene silencing and CRISPR/Cas9-gene editing) that tested OsCCA1 and OsPRR1 function. Interesting, tiller-bud outgrowth increased or decreased when OsCCA1 was either down- or up-regulated, respectively; altering OsPRR1 levels produced opposite effects. SL is a known inhibitor of rice tillering (Wang et al. 2018). To connect tiller outgrowth with OsCCA1 function, the authors mined published chromatin immunoprecipitation sequencing data for Arabidopsis CCA1 and panned for genes in the SL pathway. Among the putative AtCCA1-SL pathway targets were rice homologs of TEOSINTE BRANCHED1 (OsTB1), DWARF10 (D10), D14 and IDEAL PLANT ARCHITECTURE1 (IPA1), all of which house CCA1-binding sites in their respective promoters.
OsTB1 is a known repressor of tiller outgrowth (Wang et al. 2018). An elegant series of in vitro and in vivo experiments demonstrated that OsTB1 is a direct target of OsCCA1. As an added brake on tiller outgrowth, the SL-biosynthesis gene DWARF 10 (D10) was also shown to be a direct target of OsCCA1. Genetic interaction and overexpression analyses placed OsTB1, D10, and SL signal-encoding genes D14 and D53 downstream of OsCCA1. The authors then probed for factor(s) operating upstream of OsCCA1.
Sugars promote axillary bud outgrowth in pea (Wang et al. 2018) and influence CCA1 levels through PRR7 in Arabidopsis (Haydon et al. 2013). The authors hypothesized that sugar regulates OsCCA1 levels, which would in turn impact tillering. To test this hypothesis, a series of sugar induction experiments were conducted in rice shoots that utilized endosperm removal and exogenous sugar supplementation, panicle removal, and chemical inhibition of photosynthesis. OsCCA1 and OsTB1 expression were consistently downregulated in tiller buds, and subsequently, tiller outgrowth increased, supporting the idea that sugar, in part, governs the clock to regulate tiller bud dormancy release.
Among the putative OsCCA1 targets was IDEAL PLANT ARCHITECTURE1 (IPA1), an integral regulator of inflorescence architecture and yield in rice. The authors found that panicle size increased in OsCCA1 overexpression lines and conversely was decreased by OsCCA1 silencing. Altering OsPRR1 levels yielded opposite effects on panicle architecture. Through a strong series of in vitro and in vivo experiments, along with generating a novel knockout allele of IPA1 by CRISPR/Cas9, and performing genetic interaction and overexpression studies, IPA1 function was placed downstream of OsCCA1 and shown to be a direct target of OsCCA1.
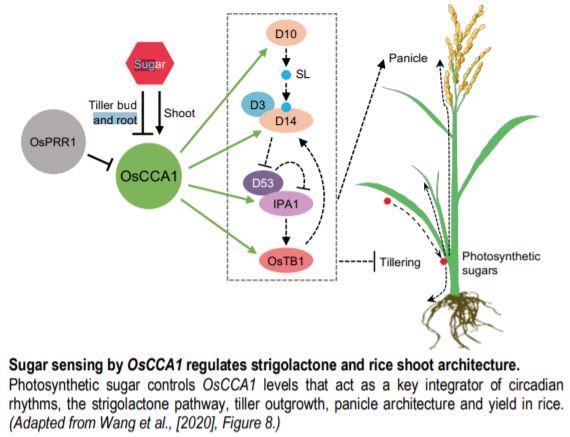
Wang and coworkers (2020) summarize their in-depth study with a model (see figure) and leave some questions to be further explored. For example, how does the clock integrate multiple factors to balance tillering and panicle development? Does IPA1 converge on similar target genes in the shoot apex and panicle? How does sugar regulate OsCCA1 expression positively in shoots but negatively in roots? Open and compelling questions raised by this important work provide several avenues for future studies on this exciting topic.
Josh Strable
Plant Biology Section
School of Integrative Plant Science
Cornell University
ORCID: 0000-0002-0260-8285
REFERENCES
Greenham, K. and McClung, C.R. (2015). Integrating circadian dynamics with physiological processes in plants. Nat. Rev. Genet. 16: 598-610.
Haydon, M.J., Mielczarek, O., Robertson, F.C., Hubbard, K.E., and Webb, A.A. (2013). Photosynthetic entrainment of the Arabidopsis thaliana circadian clock. Nature 502: 689-692.
Murakami, M., Tago, Y., Yamashino, T. and Mizuno, T. (2007). Comparative overviews of clock-associated genes of Arabidopsis thaliana and Oryza sativa. Plant Cell Physiol. 48: 110-121.
Wang, B., Smith, S.M., and Li, J. (2018). Genetic regulation of shoot architecture. Annu. Rev. Plant Biol. 69: 437-468.