Structural basis for WUSCHEL binding (bioRxiv)
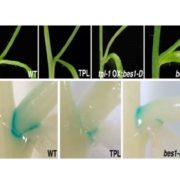
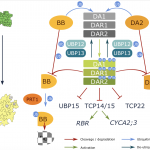
 The transcription factor WUSCHEL (WUS) plays a central role in organization of the shoot meristem. The three-helix bundle homeodomain in WUS can bind to several distinct DNA sequence motifs in many target genes promoters, but a structural view of these binding events has been lacking. Here Sloan et al. solve the structure of the WUS homeodomain (WUS-HD) bound to DNA probes containing “TAAT”, tandem “TGAA” repeats, and the G-box sequences derived from WUS-regulated genes. Overall, the WUS-HD interacts with DNA as a typical homeodomain, with conserved residues in the recognition helix inserting into the DNA major groove, but the authors also find non-conserved, WUS family-specific residues that make significant protein-DNA contacts. In contrast to many other homeodomain transcription factors, in all cases WUS most likely binds the target as a dimer. The authors then provide evidence, including cooperative binding through DNA-mediated WUS-WUS interactions and a favorable DNA helix shape, for why the direct TGAATGAA repeat is the most high-affinity target, followed by the G-box and TAAT. By comparing their results to whole genome ChiP-seq data, they further suggest that these relative binding affinities obtained in vitro actually translate to real cells. Altogether, these results provide a rational foundation for further exploration of the complex regulatory landscape of WUS-dependent transcription. (Summary by Frej Tulin) bioRxiv 10.1101/2020.03.26.009761v2
The transcription factor WUSCHEL (WUS) plays a central role in organization of the shoot meristem. The three-helix bundle homeodomain in WUS can bind to several distinct DNA sequence motifs in many target genes promoters, but a structural view of these binding events has been lacking. Here Sloan et al. solve the structure of the WUS homeodomain (WUS-HD) bound to DNA probes containing “TAAT”, tandem “TGAA” repeats, and the G-box sequences derived from WUS-regulated genes. Overall, the WUS-HD interacts with DNA as a typical homeodomain, with conserved residues in the recognition helix inserting into the DNA major groove, but the authors also find non-conserved, WUS family-specific residues that make significant protein-DNA contacts. In contrast to many other homeodomain transcription factors, in all cases WUS most likely binds the target as a dimer. The authors then provide evidence, including cooperative binding through DNA-mediated WUS-WUS interactions and a favorable DNA helix shape, for why the direct TGAATGAA repeat is the most high-affinity target, followed by the G-box and TAAT. By comparing their results to whole genome ChiP-seq data, they further suggest that these relative binding affinities obtained in vitro actually translate to real cells. Altogether, these results provide a rational foundation for further exploration of the complex regulatory landscape of WUS-dependent transcription. (Summary by Frej Tulin) bioRxiv 10.1101/2020.03.26.009761v2
[altmetric doi=”10.1101/2020.03.26.009761v2″ details=”right” float=”right”]