SNAP ‘n’ Track: Protein localization using fluorescent dyes (Plant Cell)
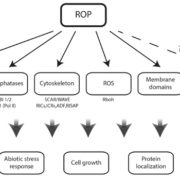
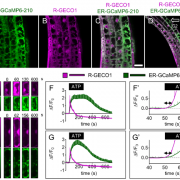
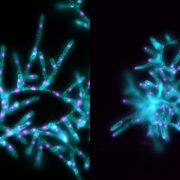
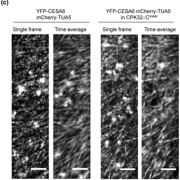
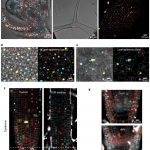
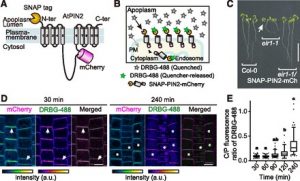 Proteins’ sub-cellular localizations provide a wealth of information regarding their functional attributes. Protein localization in plant cells is usually done through genetically combining fluorescent proteins to the protein-of-interest. Now, Iwatate and colleagues report the successful localization of target proteins in planta by adding to the protein of interest a tag that is subsequently covalently attached to a synthetic dye, in an approach termed O6-alkylguanine-DNA alkyltransferase (SNAP) tagging. The authors employed different commercial SNAP dyes to label tagged microtubules in stably transformed tobacco BY-2 cells and Arabidopsis seedlings and found them to efficiently track microtubule changes during cell division. To examine polar protein localization, the authors tracked the endocytosis of auxin efflux transporter PIN2 using DRBG-488 dye and mCherry tag and found that the dye faithfully labels PIN2 and could track its internalization. They also found that, as shown previously, freshly synthesized PIN2 proteins are preferentially deposited at the cell plate during cell division in Arabidopsis roots. This report, thus, confirms the veracity of a powerful protein tagging technique in plants. (Summary by Pavithran Narayanan @pavi_narayanan) Plant Cell 10.1105/tpc.20.00439
Proteins’ sub-cellular localizations provide a wealth of information regarding their functional attributes. Protein localization in plant cells is usually done through genetically combining fluorescent proteins to the protein-of-interest. Now, Iwatate and colleagues report the successful localization of target proteins in planta by adding to the protein of interest a tag that is subsequently covalently attached to a synthetic dye, in an approach termed O6-alkylguanine-DNA alkyltransferase (SNAP) tagging. The authors employed different commercial SNAP dyes to label tagged microtubules in stably transformed tobacco BY-2 cells and Arabidopsis seedlings and found them to efficiently track microtubule changes during cell division. To examine polar protein localization, the authors tracked the endocytosis of auxin efflux transporter PIN2 using DRBG-488 dye and mCherry tag and found that the dye faithfully labels PIN2 and could track its internalization. They also found that, as shown previously, freshly synthesized PIN2 proteins are preferentially deposited at the cell plate during cell division in Arabidopsis roots. This report, thus, confirms the veracity of a powerful protein tagging technique in plants. (Summary by Pavithran Narayanan @pavi_narayanan) Plant Cell 10.1105/tpc.20.00439