Multiple QTL mapping in autopolyploids: A random-effect model approach with application in a hexaploid sweetpotato full-sib population (Genetics)
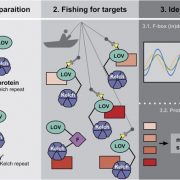
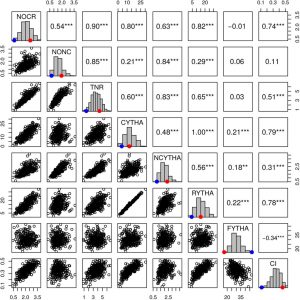 Gene mapping and QTL identification for the improvement of important traits have not been fully explored in an autopolyploid species like sweetpotatoes (2n=6x=90) due to its genetic complexity. Most sweet potato QTL mapping efforts have relied on models used for diploid species, leading to low density maps with restrictions on the statistical power for QTL detection. In tetraploids and hexaploids, the fixed-effect interval mapping (FEIM) model has also enabled easier QTL detection. Here, G. da Silva Pereira et al. explored the random-effect model (REMIM) approach and mapped multiple QTLs for yield-related traits from a sweetpotato biparental population. They compared the number of QTLs detected by the FEIM and REMIM models, and found both to be similar for yield-related traits such as storage root yield and quality and resistance to biotic and abiotic stresses, however detection power for LOD threshold was higher in the REMIM. In addition, the REMIM model was able to detect the heritability and correlation of different parental populations under varying environment and increasingly difficult scenarios. This QTL mapping method will breeding in sweetpotatoes and other autopolyploid species. (Summary by Modesta Abugu @Modestannedi) Genetics 10.1534/genetics.120.303080
Gene mapping and QTL identification for the improvement of important traits have not been fully explored in an autopolyploid species like sweetpotatoes (2n=6x=90) due to its genetic complexity. Most sweet potato QTL mapping efforts have relied on models used for diploid species, leading to low density maps with restrictions on the statistical power for QTL detection. In tetraploids and hexaploids, the fixed-effect interval mapping (FEIM) model has also enabled easier QTL detection. Here, G. da Silva Pereira et al. explored the random-effect model (REMIM) approach and mapped multiple QTLs for yield-related traits from a sweetpotato biparental population. They compared the number of QTLs detected by the FEIM and REMIM models, and found both to be similar for yield-related traits such as storage root yield and quality and resistance to biotic and abiotic stresses, however detection power for LOD threshold was higher in the REMIM. In addition, the REMIM model was able to detect the heritability and correlation of different parental populations under varying environment and increasingly difficult scenarios. This QTL mapping method will breeding in sweetpotatoes and other autopolyploid species. (Summary by Modesta Abugu @Modestannedi) Genetics 10.1534/genetics.120.303080