Robust transcriptional indicators of immune cell death revealed by spatio-temporal transcriptome analyses (Mol Plant)
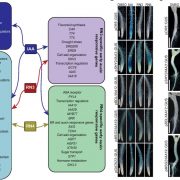
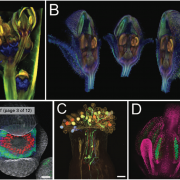
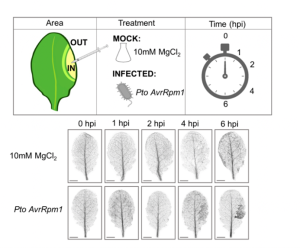 Many plants trigger a form of cell death, known as the hypersensitive response (HR), immediately upon pathogen recognition. To minimize damage to the plant, this cell death must be highly localized to the site of attack, while more distal cells survive and activate other immune responses. However, the transcriptomic programs underlying these disparate cell fates have yet to be uncovered. Here, Salguero-Linares et al. utilized an Arabidopsis thaliana–Pseudomonas syringae system and RNA-sequencing technology to isolate genes specifically implicated in hypersensitive cell death. By comparing the transcriptomes of infected areas with that of tissue outside of the inoculation zone across 5 different time points, the group were able to identify 13 marker genes for HR. The researchers then generated transgenic plants expressing green fluorescence protein under the promoters of each of the 13 genes. One line, corresponding to an uncharacterised AAA-ATPase, showed a specific fluorescence signal only in the pathogen inoculated areas destined to undergo HR, indicating this is a marker for cells destined to die. Interestingly, these plants also produced the same signals following inoculation with other bacteria which induce HR via recognition by different types of plant immune receptors, but not when inoculated with non-pathogenic bacteria, indicating that this reporter line may serve as a reliable tool for further studies of this form of plant immunity. (Summary by Rory Burke @rorby95) Mol. Plant 10.1016/j.molp.2022.04.010
Many plants trigger a form of cell death, known as the hypersensitive response (HR), immediately upon pathogen recognition. To minimize damage to the plant, this cell death must be highly localized to the site of attack, while more distal cells survive and activate other immune responses. However, the transcriptomic programs underlying these disparate cell fates have yet to be uncovered. Here, Salguero-Linares et al. utilized an Arabidopsis thaliana–Pseudomonas syringae system and RNA-sequencing technology to isolate genes specifically implicated in hypersensitive cell death. By comparing the transcriptomes of infected areas with that of tissue outside of the inoculation zone across 5 different time points, the group were able to identify 13 marker genes for HR. The researchers then generated transgenic plants expressing green fluorescence protein under the promoters of each of the 13 genes. One line, corresponding to an uncharacterised AAA-ATPase, showed a specific fluorescence signal only in the pathogen inoculated areas destined to undergo HR, indicating this is a marker for cells destined to die. Interestingly, these plants also produced the same signals following inoculation with other bacteria which induce HR via recognition by different types of plant immune receptors, but not when inoculated with non-pathogenic bacteria, indicating that this reporter line may serve as a reliable tool for further studies of this form of plant immunity. (Summary by Rory Burke @rorby95) Mol. Plant 10.1016/j.molp.2022.04.010