Ripened by Redox: Sulfoxidation of NOR Regulates Tomato Ripening
Michael J. Skelly
University of Edinburgh
Edinburgh,
United Kingdom
[email protected]
Fruit ripening is a process unique to plants that makes their fruit more appealing to seed-dispersing animals. Ripening involves a combination of physiological and metabolic reprogramming events that lead to dramatic changes in the colour, taste, texture and aroma of fruits. Tomato (Solanum lycopersicum) is an economically important crop and an excellent model organism in which to study ripening due to its simple genetics, wealth of genomic resources, ease of transformation and short life cycle (Karlova et al., 2014). Fruit ripening is underpinned by large-scale changes in gene expression, controlled by a complex network of transcriptional regulators (Giovannoni et al., 2017).
NON-RIPENING (NOR) is a NAC-domain transcription factor involved in tomato ripening. NOR has long been considered a master regulator of ripening due to the severe non-ripening phenotype of nor mutants. Recent re-evaluation of natural mutations using genome editing technology has revealed that the nor mutation is a gain-of-function allele, with a frameshift mutation resulting in a dominant-negative NOR protein (Wang et al., 2019; Wang et al., 2020). Loss-of-function nor mutants were subsequently shown to have a milder phenotype than the dominant-negative mutant (Wang et al., 2019). Nonetheless, it is clear that NOR plays a central role in tomato ripening and while its downstream effects are well established, how the activity of NOR itself might be regulated remains obscure.
Transcription factors are often regulated by post-translation modifications in response to developmental or environmental cues (Skelly et al., 2016). Oxidation of sulfur-containing amino acid residues within proteins can act as reversible redox-based switches to propagate cellular signals. While cysteine residues are well-established as targets of redox modifications (Bak et al., 2019), methionine residues are also emerging as targets of reversible modification (Rey and Tarrago, 2018). Methionine sulfoxidation can occur under oxidative conditions and is reversed by methionine sulfoxide reductase (Msr) enzymes.
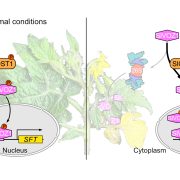
 In this issue of Plant Physiology, Jiang et al. (2020) report that NOR is regulated by Msr activity to influence tomato ripening. The authors showed that during ripening, hydrogen peroxide levels and global protein oxidation increase, suggesting that modification of protein redox status is related to ripening. By examining the expression of all Msr genes during ripening, they observed that transcripts of two of them, E4 and SlMsrB2, accumulated in fruit tissue to a much greater level than those of any other Msr genes. Furthermore, E4 and SlMsrB2 showed a similar expression pattern to NOR, increasing as ripening progressed. Using various techniques, Jiang et al. (2020) demonstrated that NOR interacts with both E4 and SlMsrB2 in vitro and in vivo. Together, these results suggest that NOR might be a substrate of Msr enzymes during tomato ripening.
In this issue of Plant Physiology, Jiang et al. (2020) report that NOR is regulated by Msr activity to influence tomato ripening. The authors showed that during ripening, hydrogen peroxide levels and global protein oxidation increase, suggesting that modification of protein redox status is related to ripening. By examining the expression of all Msr genes during ripening, they observed that transcripts of two of them, E4 and SlMsrB2, accumulated in fruit tissue to a much greater level than those of any other Msr genes. Furthermore, E4 and SlMsrB2 showed a similar expression pattern to NOR, increasing as ripening progressed. Using various techniques, Jiang et al. (2020) demonstrated that NOR interacts with both E4 and SlMsrB2 in vitro and in vivo. Together, these results suggest that NOR might be a substrate of Msr enzymes during tomato ripening.
To test whether NOR is indeed a Msr substrate, the authors oxidized purified recombinant NOR before incubating it with recombinant E4 or SlMsrB2. Both Msr enzymes were capable of reducing oxidized NOR, and mass spectrometry analysis identified four methionine residues that might be targets of sulfoxidation. The authors also examined the redox status of NOR in tomato fruit during ripening. They generated transgenic p35S:NOR-GFP plants and performed pull-down assays coupled to mass spectrometry. These experiments revealed that the methionine at position 138 (Met-138) appears to be the sole site of sulfoxidation in vivo.
The identification of NOR as a potential substrate of E4 and SlMsrB2 begs the question of how the methionine redox status of NOR affects its function as a transcription factor. To answer this, the authors generated a mutant NOR in which Met-138 was replaced with a glutamine to mimic sulfoxidation (NOR-M138Q). This mutant NOR as well as oxidized wild-type NOR were compromised in binding to target DNA sequences in vitro, as shown by electrophoretic mobility shift assays. Furthermore, NOR-M138Q was defective in transcriptional activation of target gene sequences, demonstrated by transient dual luciferase assays in Nicotiana benthamiana. Together, these data suggest that sulfoxidation of NOR at Met-138 inhibits its DNA binding and transcriptional activity. The authors analysed the protein sequence surrounding NOR Met-138 and observed that identical motifs were conserved in different NAC transcription factors from a wide range of plant species. They made mutants mimicking methionine sulfoxidation in five of these other transcription factors, and transcriptional activities were inhibited for all proteins tested. This suggests that methionine sulfoxidation of NAC transcription factors might be conserved across the plant kingdom as a mechanism to regulate transcriptional activation.
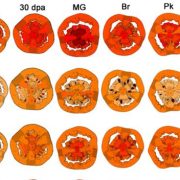
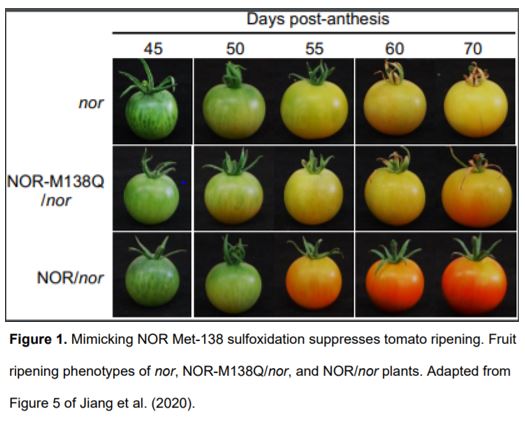
To uncover the effect of NOR methionine sulfoxidation in vivo, the authors generated transgenic plants expressing either NOR or NOR-M138Q in the nor mutant background. Plants expressing NOR (NOR/nor) exhibited almost full rescue of the ripening defects of nor, while plants expressing NOR-M138Q (NOR-M138Q/nor) showed only partial rescue (Fig. 1). These results suggest that methionine sulfoxidation of NOR inhibits tomato ripening. The authors also performed RNA-sequencing to compare the transcriptomes of these transgenic plants. Compared to nor plants, genes were up- or downregulated to a much greater extent in the fruit of NOR/nor plants than NOR-M138Q/nor plants, suggesting that mimicking sulfoxidation of NOR compromises its ability to activate target gene expression during ripening. To further test this hypothesis, the authors performed chromatin-immunoprecipitation assays to examine NOR binding at ripening-related gene promoters. These experiments showed that mimicking sulfoxidation at Met-138 decreased NOR binding at target gene promoters. Additionally, the authors showed that NOR directly binds the promoters of both E4 and SlMsrB2 and activates their expression, suggesting a potential mechanism by which NOR might regulate its own activity through a feed-forward signalling loop.
In summary, this exciting work provides evidence for post-translational regulation of NOR and uncovers a role for methionine sulfoxidation in regulating tomato ripening. The authors propose a model in which E4 and SlMsrB2 accumulate throughout ripening to maintain NOR activity in an increasingly oxidative cellular environment. Redox signalling appears to be important for fruit ripening (Decros et al., 2019), but until now, no roles for methionine sulfoxidation have been reported. This work by Jiang et al. (2020) provides new insights into not only tomato ripening, but also how transcription factors in general might be regulated by methionine sulfoxidation. Future work should focus on how NOR sulfoxidation can be engineered to alter ripening in tomato and perhaps in other fruits, with potential for profound economic and societal impacts in the agricultural and food industries.
LITERATURE CITED
Bak DW, Bechtel TJ, Falco JA, Weerapana E (2019) Cysteine reactivity across the subcellular universe. Curr Opin Chem Biol 48: 96-105
Decros G, Baldet P, Beauvoit B, Stevens R, Flandin A, Colombie S, Gibon Y, Petriacq P (2019) Get the Balance Right: ROS Homeostasis and Redox Signalling in Fruit. Front Plant Sci 10: 1091
Giovannoni J, Nguyen C, Ampofo B, Zhong S, Fei Z (2017) The Epigenome and Transcriptional Dynamics of Fruit Ripening. Annu Rev Plant Biol 68: 61-84
Karlova R, Chapman N, David K, Angenent GC, Seymour GB, de Maagd RA (2014) Transcriptional control of fleshy fruit development and ripening. J Exp Bot 65: 4527-4541
Rey P, Tarrago L (2018) Physiological Roles of Plant Methionine Sulfoxide Reductases in Redox Homeostasis and Signaling. Antioxidants (Basel) 7
Skelly MJ, Frungillo L, Spoel SH (2016) Transcriptional regulation by complex interplay between post-translational modifications. Curr Opin Plant Biol 33: 126-132
Wang R, Angenent GC, Seymour G, de Maagd RA (2020) Revisiting the Role of Master Regulators in Tomato Ripening. Trends Plant Sci 25: 291-301
Wang R, Tavano E, Lammers M, Martinelli AP, Angenent GC, de Maagd RA (2019) Re-evaluation of transcription factor function in tomato fruit development and ripening with CRISPR/Cas9-mutagenesis. Sci Rep 9: 1696