Review: Ten years of the maize nested association mapping population: Impact, limitations, and future directions (Plant Cell)
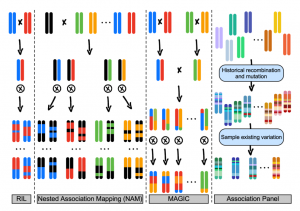 Linking phenotype to genotype is a major obstacle in plant biology, and conventional approaches (linkage analysis, association mapping) have limitations. Here Gage et al. review the past, present, and future impacts of a maize Nested Association Mapping (NAM) population developed throughout the 2000s. Designed to capitalize on the advantages while overcoming the weaknesses of traditional approaches, NAM is more powerful for detecting causal loci. The maize NAM population consists of biparental crosses conducted using the common parent B73 and 25 founder lines. After several rounds of selfing, 200 progeny were used per cross, resulting in a population of 5,000 individuals. In 2009 the population was made publicly available, representing an invaluable resource for maize researchers. Widely used for several purposes, NAM particularly facilitated identifying the genetic architecture of complex traits in maize. For example, the majority of phenotypic variation for flowering time was found to be controlled by numerous small effect loci, contrasting with observations from Arabidopsis and rice. Furthermore, from this NAM arose extensive, publicly available maize phenotyping data, and similar NAM-style approaches were incited in other species, including rice, wheat, and soybean. The synergistic, collaborative use of NAM amongst the maize community demonstrates the substantial success achievable when open science is embraced. With genome assemblies for the NAM parents recently released, the maize NAM population will advance crop improvement in the coming decade and beyond. (Summary by Caroline Dowling @CarolineD0wling) Plant Cell 10.1105/tpc.19.00951
Linking phenotype to genotype is a major obstacle in plant biology, and conventional approaches (linkage analysis, association mapping) have limitations. Here Gage et al. review the past, present, and future impacts of a maize Nested Association Mapping (NAM) population developed throughout the 2000s. Designed to capitalize on the advantages while overcoming the weaknesses of traditional approaches, NAM is more powerful for detecting causal loci. The maize NAM population consists of biparental crosses conducted using the common parent B73 and 25 founder lines. After several rounds of selfing, 200 progeny were used per cross, resulting in a population of 5,000 individuals. In 2009 the population was made publicly available, representing an invaluable resource for maize researchers. Widely used for several purposes, NAM particularly facilitated identifying the genetic architecture of complex traits in maize. For example, the majority of phenotypic variation for flowering time was found to be controlled by numerous small effect loci, contrasting with observations from Arabidopsis and rice. Furthermore, from this NAM arose extensive, publicly available maize phenotyping data, and similar NAM-style approaches were incited in other species, including rice, wheat, and soybean. The synergistic, collaborative use of NAM amongst the maize community demonstrates the substantial success achievable when open science is embraced. With genome assemblies for the NAM parents recently released, the maize NAM population will advance crop improvement in the coming decade and beyond. (Summary by Caroline Dowling @CarolineD0wling) Plant Cell 10.1105/tpc.19.00951