Revealing the Invisible: A Synthetic Reporter for ABA
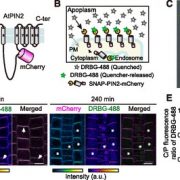
How do you make the invisible visible? It’s a question that crops up again and again in molecular biology. Abscisic acid (ABA) is one of the most important hormones in plants and is essential for survival in suboptimal conditions. Despite this, we have only a basic understanding of where and when this hormone acts. ABA can be measured directly from homogenized samples, but this approach is mainly restricted to organ-level analysis (McAdam, 2015). Recently, two reporter systems were developed that used Förster resonance energy transfer to visualize ABA concentrations in intact plants (Jones et al., 2014; Waadt et al., 2014). Although it is likely that these reporters will improve in future generations, they are currently not sensitive enough to capture endogenous ABA at a tissue-level resolution.
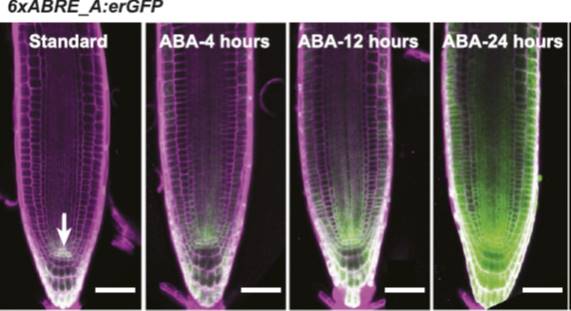
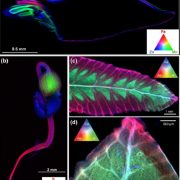
In this issue of Plant Physiology, Wu et al. (2018) have taken a different approach. Rather than try to visualize ABA levels directly, they instead produced a synthetic transcriptional reporter of ABA. It has long been known that many of the genes that are regulated by ABA have a specific sequence motif (ACGTGTC) in their promoters, known as the ABA-Responsive Element or ABRE (Mundy et al., 1990). Wu et al. used this knowledge to create a synthetic promoter, composed of six repeats of the ABRE (6xABRE), akin to the widely used synthetic auxin-responsive promoter DR5 (Ulmasov et al., 1997). The 6xABRE promoter was then fused to an endoplasmic reticulum-localized GFP. The resulting reporter allows for the visualization of ABA-induced transcriptional activity at the cellular level.
In the presence of ABA, several bZIP transcription factors show enhanced binding to ABREs (Song et al., 2016). Accordingly, the 6xABRE-reporter was strongly induced by exogenous ABA in the root tip, lateral roots, and stomata. Importantly, the reporter was also induced by salinity and osmotic stress, which enhance the production of endogenous ABA in plants. By varying the number of ABRE repeats and the sequences flanking the ABRE within the repeats, the authors found that they could alter the inducibility of their reporter. The three bases immediately following the ABRE play a particularly important role in dictating which transcription factors will bind to the element.
Unexpectedly, the group found very high 6xABRE reporter activity in the quiescent center of the root, but this expression appeared to be independent of ABA signaling. In an effort to discover how this happens, they conducted large-scale yeast one-hybrid screen to see which transcription factors bind to the 6xABRE promoter. In addition to the expected members of the bZIP transcription factor family, the group found that the meristem specification factor WUSCHEL (WUS) and a regulator of cell death, NAC13, also bind to the promoter. A close relative of WUS, WOX5, promoted and NAC13 repressed the expression of the 6xABRE reporter in the quiescent center, indicating that the ABRE element can be regulated by transcription factors outside of the bZIP family, in an ABA-independent manner.
The finding that the 6xABRE reporter was activated in certain cell types independently of ABA demonstrates some of the inherent limitations of transcription-based reporters for measuring hormone activity. Because they act a long way downstream of hormone perception, it is always possible that reporter expression is controlled by transcription factors that are not related to the hormone being studied.
It may, however, be possible to overcome some of these problems, first by always using appropriate controls. Expressing the 6xABRE in an ABA signaling mutant would allow for the deduction of ABA-dependent and -independent effects. Second, it may be possible to alter the sequence of the DNA binding element or flanking regions to improve the specificity of the reporter. Another approach would be to express a transcriptional reporter alongside a more direct measure of hormone levels (such as the Förster resonance energy transfer sensors described previously) to track both hormone levels and activity simultaneously. The former two approaches were recently applied to great effect in the context of auxin signaling, by expressing an improved version of the DR5 transcriptional reporter along with the more direct reporter of auxin levels, DII-Venus (Liao et al., 2015).
There will always room for improvement of hormone reporters as our knowledge of cell biology and genetics advances. Nevertheless, the 6xABRE reporter developed by Wu et al. is a much-needed tool for current research and will provide an excellent basis for the future development of synthetic ABA reporters.
REFERENCES
Jones AM, Danielson JA, Manojkumar SN, Lanquar V, Grossmann G, Frommer WB (2014) Abscisic acid dynamics in roots detected with genetically encoded FRET sensors. eLife 3: e01741
Liao CY, Smet W, Brunoud G, Yoshida S, Vernoux T, Weijers D (2015) Reporters for sensitive and quantitative measurement of auxin response. Nat Methods 12: 207–210
McAdam S (2015) Physicochemical quantification of abscisic acid levels in plant tissues with an added internal standard by ultra-performance liquid chromatography. Bio Protoc 5: e1599
Mundy J, Yamaguchi-Shinozaki K, Chua NH (1990) Nuclear proteins bind conserved elements in the abscisic acid-responsive promoter of a rice rab gene. Proc Natl Acad Sci USA 87: 1406–1410
Song L, Huang SC, Wise A, Castanon R, Nery JR, Chen H, Watanabe M, Thomas J, Bar-Joseph Z, Ecker JR (2016) A transcription factor hierarchy defines an environmental stress response network. Science 354: aag1550
Ulmasov T, Murfett J, Hagen G, Guilfoyle TJ (1997) Aux/IAA proteins repress expression of reporter genes containing natural and highly active synthetic auxin response elements. Plant Cell 9: 1963–1971
Waadt R, Hitomi K, Nishimura N, Hitomi C, Adams SR, Getzoff ED, Schroeder JI (2014) FRET-based reporters for the direct visualization of abscisic acid concentration changes and distribution in Arabidopsis. eLife 3: e01739
Wu R, Duan L, Pruneda-Paz J, Oh D, Pound M, Kay S, Dinneny JR (2018) The 6xABRE synthetic promoter enables the spatiotemporal analysis of ABA-mediated transcriptional regulation. Plant Physiol177:1650–1665