Part of the Stack; How Does a Protein Know its Place?
Parsons et al. successfully separate early to late Golgi cisternae to reveal the sequential localization of resident proteins and the sequence features that guide transmembrane proteins within the Golgi stack. Plant Cell https://doi.org/10.1105/tpc.19.00081
By Harriet T. Parsons (University of Cambridge) and Tim J. Stevens (MRC Laboratory of Molecular Biology)
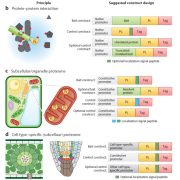
Background: The Golgi apparatus is often likened to the central sorting office of the cell. It receives material, modifies it according to requirements, then distributes it elsewhere. The Golgi manages to perform many different types of modifications on a wide range of material that flows through it, such that the correct end-product leaves the Golgi on time, destined for the correct place. The architecture of the Golgi is likely to play a key role in enabling such complex organization. The Golgi resembles a stack of pancakes. As material flows from one end of the pancake stack to the other, it is modified, stamped and addressed. In each ‘pancake’, a different set of modifications are thought to occur; each contains a unique suite of machinery (or proteins) and so performs unique chemistry.
Question: We set out to discover how the Golgi knows which protein to assign to which ‘pancake’. No one has managed to separate this pancake stack—so no one knows what each part contains, let alone how the correct machinery ends up in the correct place.
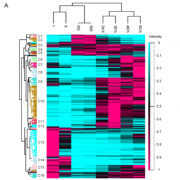
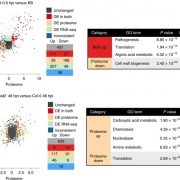
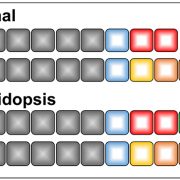
Findings: We removed Golgi stacks from cultured Arabidopsis cells by breaking the cells open under a gentle vacuum. We separated the Golgi pancakes (or cisternae) by passing a voltage across the Golgi stacks, as the cisternae at the far end of the Golgi stacks seem to be more negatively charged than those at the near end. We identified all the proteins in the entire range of separated cisternae. By seeing when the amount of each protein increased or decreased along this range, we could work out which cisternae it was in. Then we compared the detailed compositions of proteins from different cisternae. We saw distinct features depending on whether the protein was in the early, middle or late parts of the stack.
Next steps: We think these features define which pancake a protein is allowed in, and which it’s excluded form. Next we want to test the relative importance of each signature. We also want to know whether the same signatures are found in humans as well as plants.
H.T. Parsons, T. J. Stevens, H. E. McFarlane, S. Vidal-Melgosa, J. Griss, N. Lawrence, R. Butler, M. M. L. Sousa, M. Salemi, W. G. T. Willats, C. J. Petzold, J. L. Heazlewood, K. S. Lilley. (2019). Separating Golgi Proteins from Cis to Trans Reveals Underlying Properties of Cisternal Localization. Plant Cell. https://doi.org/10.1105/tpc.19.00081.
Key words: Golgi, proteomics, protein sorting