A chloroplast protein atlas reveals punctate structures and spatial organization of biosynthetic pathways
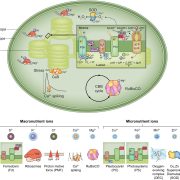
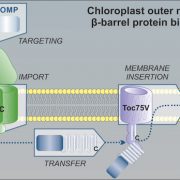
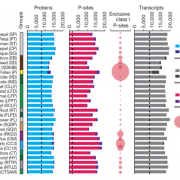
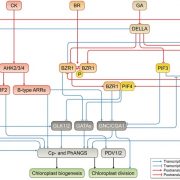
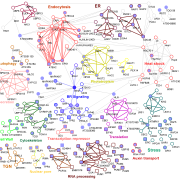
 Chloroplasts are the location of key processes including photosynthesis, starch synthesis and lipid synthesis. However, many chloroplast proteins have unknown functions, a problem that can in part by addressed through high-resolution localization data. Here, 1034 putative chloroplast- localized proteins were tagged with a C-terminal fluorescent Venus tag, constitutively expressed in the green alga Chlamydomonas reinhardtii, followed by protein localization using confocal microscopy. 28 proteins with known localizations were used to trial the method. Of these, 27 localizations matched previous results, suggesting this method has a high accuracy. Of the 1034 proteins, 581 localized to distinct chloroplast subcompartments, with the pattern of localization often providing a clue to protein function. Interestingly, some enzymes, including those in the chlorophyll biosynthetic pathways, localized to puncta that do not correspond to any chloroplast structure. These puncta might concentrate an enzyme to where its substrate is plentiful or regulate a specific reaction. If you are wondering about the localization of your favorite protein, the authors made their localization data available online and also integrated it into a protein prediction program called PB-Chlamy, both valuable resources to the wider community. (Summary by Rose McNelly @Rose_McN) Cell 10.1016/j.cell.2023.06.008
Chloroplasts are the location of key processes including photosynthesis, starch synthesis and lipid synthesis. However, many chloroplast proteins have unknown functions, a problem that can in part by addressed through high-resolution localization data. Here, 1034 putative chloroplast- localized proteins were tagged with a C-terminal fluorescent Venus tag, constitutively expressed in the green alga Chlamydomonas reinhardtii, followed by protein localization using confocal microscopy. 28 proteins with known localizations were used to trial the method. Of these, 27 localizations matched previous results, suggesting this method has a high accuracy. Of the 1034 proteins, 581 localized to distinct chloroplast subcompartments, with the pattern of localization often providing a clue to protein function. Interestingly, some enzymes, including those in the chlorophyll biosynthetic pathways, localized to puncta that do not correspond to any chloroplast structure. These puncta might concentrate an enzyme to where its substrate is plentiful or regulate a specific reaction. If you are wondering about the localization of your favorite protein, the authors made their localization data available online and also integrated it into a protein prediction program called PB-Chlamy, both valuable resources to the wider community. (Summary by Rose McNelly @Rose_McN) Cell 10.1016/j.cell.2023.06.008