Purple is the New Orange: Anthocyanin Regulation Coming Together in Carrot
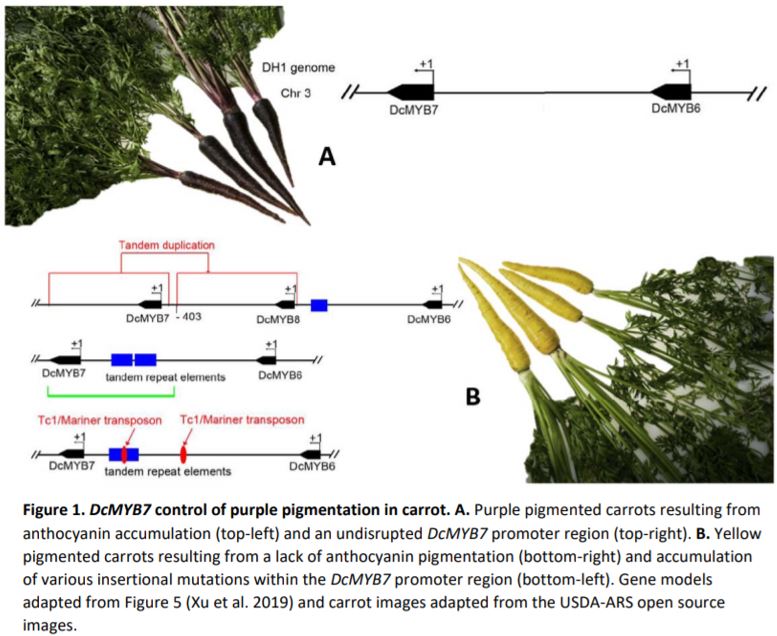
Western cultivated carrot (Daucus carota subsp. sativus L) is broadly known for its orange pigmentation and accumulation of carotenoids, known as the carotene group (var. sativus). However, the eastern wild carrot (subsp. carota) originating from southwestern Asia over a century ago provides purple pigmentation (Figure 1A). Purple carrots, known as the anthocyanin group (var. atrorubens Alef.) accumulate anthocyanins in root tissues, in addition to flowers, fruits and petioles (Kammerer et al., 2004; Arscott and Tanumihardjo, 2010; Montilla et al., 2011). Genetic control of anthocyanin accumulation in carrot has been tracked down to two separate loci, P1 and P3 on chromosome 3. The P3 locus has been shown to control solid purple pigmentation in roots (purple periderm, phloem and xylem) and petioles, but neither P1 nor P3 loci co-localize with anthocyanin biosynthetic genes. For this reason, it is assumed regulatory factors control purple pigmentation variation in wild carrot, and mutations within these genes contributed to carrot domestication—going from purple to yellow carrots, and later orange.
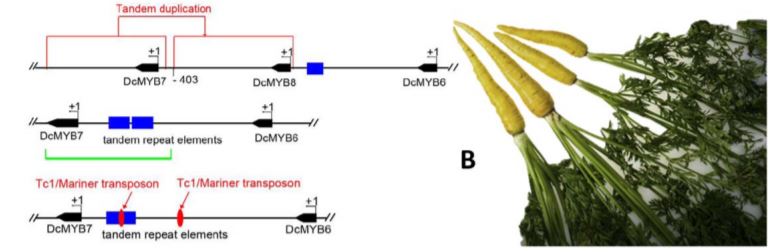
In this issue of Plant Physiology, Xu et al. 2019 made significant advancements in the characterization of two candidate genes, DcMYB6 (DCAR_000385) and DcMYB7 (DCAR_010745) which underly the P3 locus. DcMYB6 encodes a R2R3-MYB (MYB) transcription factor, a family of proteins known to interact with basic helix-loop-helix (bHLH) and WD-repeat proteins to regulate anthocyanin biosynthetic genes (Espley et al., 2007; Chagne et al., 2013; Jin et al., 2016). MYBs comprise a dynamic group of genes that cluster together within loci as MYB-like genes. The P3 locus is no different, hosting six different MYB-like genes including DcMYB6 and DcMYB7 (Figure 1A). Differential expression analysis revealed DcMYB6 as having the highest expression in purple pigmented tissues, but still detectable in non-pigmented tissues, thus making it a strong candidate for controlling anthocyanin accumulation. DcMYB7 was also expressed but at lower levels in purple tissues and was non-detectable in non-purple tissues.
Previous work from the authors showed the more highly expressed DcMYB6 was capable of stimulating anthocyanin biosynthesis in Arabidopsis (Xu et al., 2017). Nevertheless, overexpression of DcMYB6 in carrot failed to result in pigmentation during transformation or in whole plants, weakening the likely role of DcMYB6 as a major regulatory factor. In contrast, overexpression of DcMYB7 resulted in anthocyanin accumulation in both Arabidopsis and carrot, with deep purple pigmentation throughout the carrot root tissue profile, petioles and seeds, but not in petals or stamens. Further investigation of DmMYB7 was conducted by characterizing knock-out mutants generated in a solid purple carrot background. DmMYB7 knock-out mutants displayed almost complete purple depigmentation, revealing yellow pigmentation from existing lutein accumulation and confirming DcMYB7 as being a major regulatory factor in anthocyanin accumulation (Figure 1B).
These experiments demonstrated the function of DcMYB7 in anthocyanin accumulation but did not explain the variation in purple pigmentation seen across varieties. A third MYB-like gene, named DcMYB8 (DCAR_010746) helped in this explanation by being discovered within the promoter region of DcMYB7 in some non-purple varieties. Further unearthing of tandem repeat elements and a Tc1/Mariner transposon within the same promoter region of DcMYB7 in other non-purple varieties further supported the importance of DcMYB7 promoter function for anthocyanin accumulation in purple varieties, and the likely role of these accumulated mutations in the domestication of purple carrots to yellow (Figure 1B).
 The final piece of this pigmentation puzzle came from the identification of the downstream targets of DcMYB7 which influence anthocyanin biosynthesis. Anthocyanins are glycosylated and acylated for improved stability in plant tissues and storage (Kammerer et al., 2004; Montilla et al., 2011; Cavagnaro et al., 2014). The anthocyanin profiles of wild-type solid purple carrot roots versus DcMYB7 overexpression lines in the non-purple carrot background revealed an accumulation of highly glycosylated and acylated species (called Cy3SGG) in the overexpression lines. Two enzymes, UGT79B1 (At5g54060) and SAT1 (At2g23000) have been implicated in Cy3SGG accumulation in Arabidopsis (Yonekura-Sakakibara et al., 2012). Indeed, orthologs of UGT79B1 and SAT1, DcUCGXT1 and DcSAT1 respectively were upregulated in solid purple carrot varieties and even more so in DcMYB7 overexpression lines compared to non-purple carrot controls and DcMYB7 knock-out lines. Binding assays showed that DcMYB7 binds to the promoters of DcUCGXT1 and DcSAT1, indicating that they are its direct targets, and pointing to how the expression of DcMYB7 affects anthocyanin accumulation.
The final piece of this pigmentation puzzle came from the identification of the downstream targets of DcMYB7 which influence anthocyanin biosynthesis. Anthocyanins are glycosylated and acylated for improved stability in plant tissues and storage (Kammerer et al., 2004; Montilla et al., 2011; Cavagnaro et al., 2014). The anthocyanin profiles of wild-type solid purple carrot roots versus DcMYB7 overexpression lines in the non-purple carrot background revealed an accumulation of highly glycosylated and acylated species (called Cy3SGG) in the overexpression lines. Two enzymes, UGT79B1 (At5g54060) and SAT1 (At2g23000) have been implicated in Cy3SGG accumulation in Arabidopsis (Yonekura-Sakakibara et al., 2012). Indeed, orthologs of UGT79B1 and SAT1, DcUCGXT1 and DcSAT1 respectively were upregulated in solid purple carrot varieties and even more so in DcMYB7 overexpression lines compared to non-purple carrot controls and DcMYB7 knock-out lines. Binding assays showed that DcMYB7 binds to the promoters of DcUCGXT1 and DcSAT1, indicating that they are its direct targets, and pointing to how the expression of DcMYB7 affects anthocyanin accumulation.
Purple carrots are a wild throwback to the origins of carrot domestication and provide important health benefits to humans (Netzel et al., 2007; He and Giusti, 2010). Xu et al. 2019 have provided compelling insights towards understanding the genetic controls of anthocyanin accumulation in carrots which can be further applied to advance breeding and genetic efforts in carrot, and the spectrum of other purple pigmented crop species.
Nathaniel Butler, Department of Horticulture, University of Wisconsin-Madison
ORCID ID: 0000-0002-9235-7626
Literature Cited
Arscott SA, Tanumihardjo SA (2010) Carrots of many colors provide basic nutrition and bioavailable phytochemicals acting as a functional food. Compr Rev Food Sci 9: 223-239
Cavagnaro PF, Iorizzo M, Yildiz M, Senalik D, Parsons J, Ellison S, Simon PW (2014) A gene-derived SNP-based high resolution linkage map of carrot including the location of QTL conditioning root and leaf anthocyanin pigmentation. BMC Genomics 15: 1118
Chagne D, Lin-Wang K, Espley RV, Volz RK, How NM, Rouse S, Brendolise C, Carlisle CM, Kumar S, De Silva N, Micheletti D, McGhie T, Crowhurst RN, Storey RD, Velasco R, Hellens RP, Gardiner SE, Allan AC (2013) An ancient duplication of apple MYB transcription factors is responsible for novel red fruit-flesh phenotypes. Plant Physiol 161: 225-239
Espley RV, Hellens RP, Putterill J, Stevenson DE, Kutty‐Amma S, Allan AC (2007) Red colouration in apple fruit is due to the activity of the MYB transcription factor, MdMYB10. Plant J 49: 414-427
He J, Giusti MM (2010) Anthocyanins: natural colorants with health-promoting properties. Annu Rev Food Sci Technol 1: 163-187
Jin W, Wang H, Li M, Wang J, Yang Y, Zhang X, Yan G, Zhang H, Liu J, Zhang K (2016) The R2R3 MYB transcription factor PavMYB10.1 involves in anthocyanin biosynthesis and determines fruit skin colour in sweet cherry (Prunus avium L.). Plant Biotechnol J 14: 2120-2133
Kammerer D, Carle R, Schieber A (2004) Quantification of anthocyanins in black carrot extracts (Daucus carota ssp sativus var. atrorubens Alef.) and evaluation of their color properties. Eur Food Res Technol 219: 479-486
Montilla EC, Arzaba MR, Hillebrand S, Winterhalter P (2011) Anthocyanin composition of black carrot (Daucus carota ssp. sativus var. atrorubens Alef.) cultivars Antonina, Beta Sweet, Deep Purple, and Purple Haze. J Agric Food Chem 59: 3385-3390
Netzel M, Netzel G, Kammerer DR, Schieber A, Carle R, Simons L, Bitsch I, Bitsch R, Konczak L (2007) Cancer cell antiproliferation activity and metabolism of black carrot anthocyanins. Innov Food Sci Emerg 8: 365-372
Xu ZS, Feng K, Que F, Wang F, Xiong AS (2017) A MYB transcription factor, DcMYB6, is involved in regulating anthocyanin biosynthesis in purple carrot taproots. Sci Rep 7: 45324
Xu ZS, Yang QQ, Feng K, Xiong AS (2019) DcMYB7 regulates anthocyanin modification by activating DcUCGXT1 and DcSAT1 in carrot. Plant Physiol (this issue)
Yonekura-Sakakibara K, Fukushima A, Nakabayashi R, Hanada K, Matsuda F, Sugawara S, Inoue E, Kuromori T, Ito T, Shinozaki K, Wangwattana B, Yamazaki M, Saito K (2012) Two glycosyltransferases involved in anthocyanin modification delineated by transcriptome independent component analysis in Arabidopsis thaliana. Plant J 69: 154-167