Transcriptome Dynamics and Gene Activities during Embryo and Grain Development in Wheat
Xiang et al. elucidate transcriptome dynamics and gene activities during embryogenesis and grain development in modern wheat and its progenitors. Plant Cell https://doi.org/10.1105/tpc.19.00397.
By Daoquan Xiang, Teagen D. Quilichini, Peng Gao, and Raju Datla
Aquatic and Crop Resource Development, National Research Council Canada, 110 Gymnasium Place, Saskatoon, SK, S7N 0W9, Canada
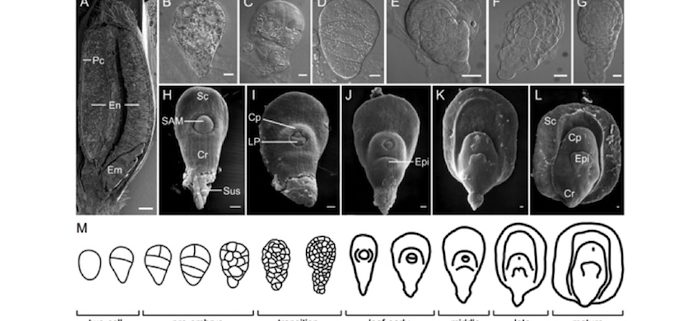
Background: Wheat is one of the most important staple global food crops and is intimately connected to human civilization. Modern wheat cultivation relies upon two polyploid species, Triticum (T.) aestivum and T. turgidum, which are putatively derived from three diploid ancestral species, T. monococcum, Aegilops (Ae) speltoides, and Ae. tauschii. The grain is the key economic product of this crop, with 20% of the calories and protein consumed by humans provided by its two predominant components, the embryo and endosperm. To explore the developmental programs of grain production in cultivated wheat and its ancestors, we performed global gene expression profiling. The aim of this study was to reveal the underpinning gene activities involved in the making of the grain in modern cultivated wheats in relation to diploid grass progenitors.
Question: We wanted to know how domestication and selective breeding has shaped the programming of gene activities during grain development in the complex polyploid genomes of cultivated wheat, the relative contributions of progenitor diploid genomes, and the effect of polyploidization on gene expression during grain development.
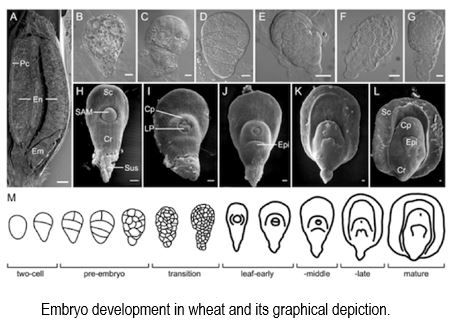 Findings: Despite sharing similarities, grain development in modern wheat cultivars displays distinct features from that of their progenitor diploid species. These differences are reflected in the programing of the diploid A, B, and D sub-genomes, which modulates their respective gene activities qualitatively and quantitatively to produce distinct gene expression signatures. This study revealed evolutionary divergence among the sub-genomes in shaping embryo and endosperm development in polyploid wheats. Through the characterization of transcriptional programming of embryogenesis in hexaploid wheat, tetraploid wheat, and ancestral diploid grass species, our work provides important insights into the evolution of gene expression in wheat and the selective pressures placed on grain production. This study expands our knowledge of the evolution of domesticated wheat.
Findings: Despite sharing similarities, grain development in modern wheat cultivars displays distinct features from that of their progenitor diploid species. These differences are reflected in the programing of the diploid A, B, and D sub-genomes, which modulates their respective gene activities qualitatively and quantitatively to produce distinct gene expression signatures. This study revealed evolutionary divergence among the sub-genomes in shaping embryo and endosperm development in polyploid wheats. Through the characterization of transcriptional programming of embryogenesis in hexaploid wheat, tetraploid wheat, and ancestral diploid grass species, our work provides important insights into the evolution of gene expression in wheat and the selective pressures placed on grain production. This study expands our knowledge of the evolution of domesticated wheat.
Next steps: The insights gained from investigating grain development in wheat reveal the constraints and flexibilities associated with grain development and regulatory programs, which will be used to explore the potential to improve grain composition and size traits. Advancing this knowledge will be helpful for wheat cultivar development to meet global food demands.
Daoquan Xiang, Teagen D. Quilichini, Ziying Liu, Peng Gao, Youlian Pan, Qiang Li, Kirby T. Nilsen, Prakash Venglat, Eddi Esteban, Asher Pasha, Yejun Wang, Rui Wen, Zhongjuan Zhang, Zhaodong Hao, Edwin Wang, Yangdou Wei, Richard Cuthbert, Leon Kochian, Andrew Sharpe, Nicholas Provart, Dolf Weijers, C. Stewart Gillmor, Curtis Pozniak, Raju Datla (2019). The Transcriptional Landscape of Polyploid Wheats and their Diploid Ancestors during Embryogenesis and Grain Development. Plant Cell 31: xxx; DOI: https://doi.org/10.1105/tpc.19.00397
Key words: Wheat, Gene Expression, Embryogenesis, Grain Development