Circadian regulation of the transcriptome in a complex polyploid crop (PLoS Biol)
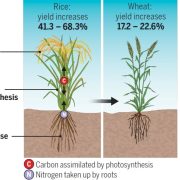
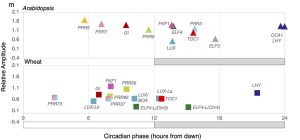 Circadian regulation fine-tunes patterns of gene expression in plants in changing environments and has been selected for during crop domestication. Many studies have been done on circadian gene expression in Arabidopsis, but far less is known about this regulation in hexaploid bread wheat, especially on how it is balanced across its three subgenomes. Here, Rees et al. present a 3-day circadian RNA-sequencing time course of aerial tissue from bread wheat (cv. Cadenza) seedlings, providing a valuable resource to explore these questions. First, they performed global analysis on the gene expression patterns and compared this to published data on Arabidopsis. Although previous work reported 72.5% balanced expression of wheat triads at a single time point, here only 11.1% were circadian balanced, which is likely due to the many criteria of circadian balance: phase, period, and rhythmicity. Overall, there were three times as many imbalanced as balanced triads in wheat, and a lower proportion of rhythmically expressed genes compared to Arabidopsis. Also, the mean period length for wheat was approx. 3 hours longer than for Arabidopsis, but there were no distinct differences between the three wheat subgenomes. Secondly, the authors compared the wheat and Arabidopsis transcriptomes for specific sets of genes. They found that the core circadian gene expression was conserved, as well as most of the circadian expression of photosynthesis, photoreceptor, and light signalling genes. However, the circadian control of primary metabolism genes was more variable, with several genes expressed rhythmically in Arabidopsis but not in wheat, or with different peak phases or relative amplitudes. (Summary by Jiawen Chen @Jiaaawen) PLoS Biol. 10.1371/journal.pbio.3001802
Circadian regulation fine-tunes patterns of gene expression in plants in changing environments and has been selected for during crop domestication. Many studies have been done on circadian gene expression in Arabidopsis, but far less is known about this regulation in hexaploid bread wheat, especially on how it is balanced across its three subgenomes. Here, Rees et al. present a 3-day circadian RNA-sequencing time course of aerial tissue from bread wheat (cv. Cadenza) seedlings, providing a valuable resource to explore these questions. First, they performed global analysis on the gene expression patterns and compared this to published data on Arabidopsis. Although previous work reported 72.5% balanced expression of wheat triads at a single time point, here only 11.1% were circadian balanced, which is likely due to the many criteria of circadian balance: phase, period, and rhythmicity. Overall, there were three times as many imbalanced as balanced triads in wheat, and a lower proportion of rhythmically expressed genes compared to Arabidopsis. Also, the mean period length for wheat was approx. 3 hours longer than for Arabidopsis, but there were no distinct differences between the three wheat subgenomes. Secondly, the authors compared the wheat and Arabidopsis transcriptomes for specific sets of genes. They found that the core circadian gene expression was conserved, as well as most of the circadian expression of photosynthesis, photoreceptor, and light signalling genes. However, the circadian control of primary metabolism genes was more variable, with several genes expressed rhythmically in Arabidopsis but not in wheat, or with different peak phases or relative amplitudes. (Summary by Jiawen Chen @Jiaaawen) PLoS Biol. 10.1371/journal.pbio.3001802