Prospects of Single Cell Transcriptomics in Plant Biology Research
In recent years, plant single-cell transcriptomics has been developing rapidly and enabling researchers to study plant organs with high resolution and high throughput information. Single cell transcriptome technology was first reported in 2009 (Tang et al.,2009) and as the name suggests, single cell transcriptome is the transcriptome of a single cell. Single cell-RNA sequencing (scRNA-sequencing) and single-nucleus RNA sequencing (snRNA-sequencing) technologies help us gain insights into each cell type, classify cell type and cell development trajectory as well as gene regulatory networks of plants (Chen et al., 2023; Zheng et al.,2023). Most of the single cell transcriptomic studies are on model plant Arabidopsis, followed by Zea mays and Oryza sativa (Zheng et al.,2023). These technologies are used depending on the choice of tissue type required by researchers which may be either cell or nuclei (Zheng et al.,2023; Seyfferth et al.,2021). Both methods have the disadvantage of losing spatial information of the cells when isolating them from tissues, thus spatial transcriptome technology has undergone considerable development in recent years, wherein spatial transcriptomics preserves spatial information while profiling the transcriptome which provides more precise results for studying gene regulatory networks (Chen et al.,2023) (Figure 1). These technologies use three main single-cell sequencing platforms: plate-based, combinatorial indexing based, and bead based out of which 10X genomics bead based method has been the most popularly used and scRNA-sequencing has been the most widely used in plants (Zheng et al.,2023).
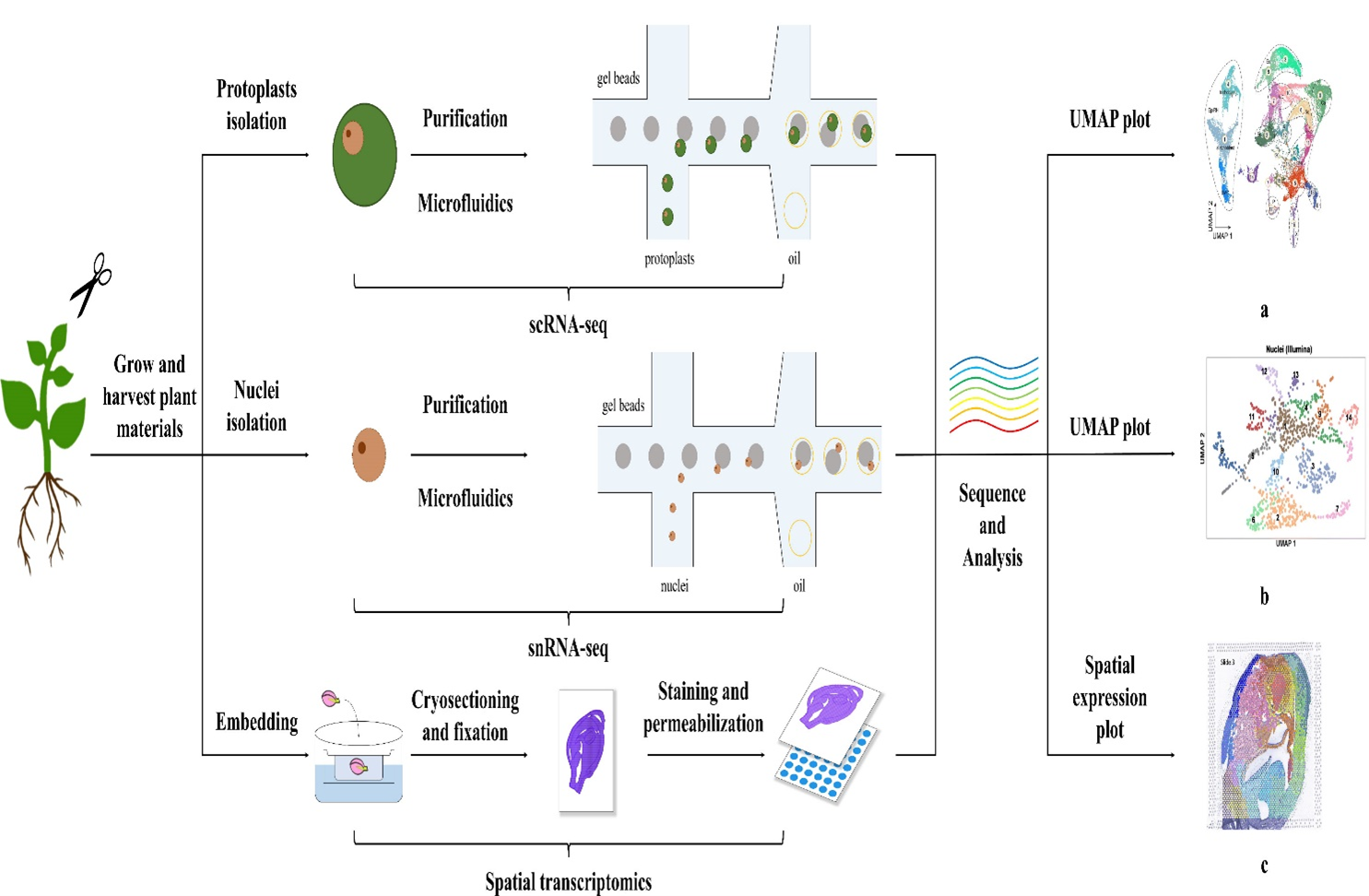
Figure 1: Illustration of high throughput sequencing technologies at single-cell resolution for plants (Source-Chen et al.,2023 Frontiers in Plant Science)
Researchers have constructed gene expressional maps for diverse tissues at single cell resolution in Arabidopsis, such as lateral roots (Serrano-Ron et al.,2021; Shahan et al.,2022); these technologies allow precise identification of developmental trajectories of identities like vascular tissues vegetative shoot apex (Zhang et al.,2021) and blade vascular system (Kim et al.,2021). scRNA-sequencing can also play a role in explaining plant hormone function with the analysis of plant transcription factors (Liu et al.,2022). Gene pathways involving metabolite biosynthesis can be explored through scRNA-sequencing (Kang et al.,2022). Although single cell transcriptomic approaches have introduced novel opportunities for plants, the coming years will be characterized by applications mentioned in Figure 2. For easier interpretation of plant single-cell RNA analysis, there are now comprehensive plant-single cell transcriptomic databases specifically like PlantscRNAdb (Chen et al.,2021), PsctH (Xu et al.,2022) and PCMDB (Jin et al.,2022). Initiatives like Plant Cell Atlas (Jha et al.,2021) are now further assisting in uniting scRNA-seq data sets and research tools for the scientific community. There is no denying that single-cell transcriptomics offer solutions to longstanding biological queries but efficient cell isolation along with unified experimental design and analysis is a must for these technologies to create ample research possibilities.
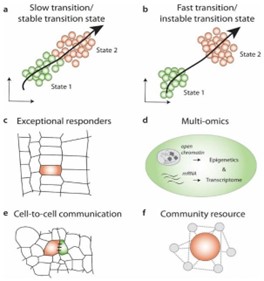
Figure 2: Application of single-cell technologies in plant cell research (a-b) Enrichment of subpopulation can be used to identify and characterize cell state changes along a developmental trajectory. Slow transitions between two states suggest gradual changes in the transcriptome, while fast cell state transitions, without intermediate stages, can suggest a switch-like behavior in the cell states. (c) Ultra high-throughput analysis of ten-to hundreds of thousands of single cells could reveal heterogeneity within cells of the same cell type. The transcriptomes of such exceptional responders carry useful information to understand phenotypic plasticity. (d) Multi-omics single cell approaches can be used to correlate cell-specific transcriptome profiles with gene regulatory elements or other cellular information (metabolome, proteome, etc.). (e) Combining single cell transcriptomic profiles with spatial information can reveal cell-to-cell communication signals as seen in ligand-receptor mediated pathways. (f) Data depository and integration initiatives, like the Plant Cell Atlas, aim to unify experimental conditions and sample processing to allow a standardized analysis and integration of scRNA-seq data sets as valuable community resources. (Source: Seyfferth et al., 2021 Annual Reviews of Plant Biology)
References:
- Tang, F., Barbacioru, C., Wang, Y., Nordman, E., Lee, C., Xu, N., … & Surani, M. A. (2009). mRNA-Seq whole-transcriptome analysis of a single cell. Nature methods, 6(5), 377-382.
- Chen, C., Ge, Y., & Lu, L. (2023). Opportunities and challenges in the application of single-cell and spatial transcriptomics in plants. Frontiers in Plant Science, 14.
- Zheng, D., Xu, J., Lu, Y., Chen, H., Chu, Q., & Fan, L. (2023). Recent progresses in plant single-cell transcriptomics. Crop Design, 100041.
- Seyfferth, C., Renema, J., Wendrich, J. R., Eekhout, T., Seurinck, R., Vandamme, N., … & De Rybel, B. (2021). Advances and opportunities in single-cell transcriptomics for plant research. Annual Review of Plant Biology, 72, 847-866.
- Serrano-Ron, L., Perez-Garcia, P., Sanchez-Corrionero, A., Gude, I., Cabrera, J., Ip, P. L., … & Moreno-Risueno, M. A. (2021). Reconstruction of lateral root formation through single-cell RNA sequencing reveals order of tissue initiation. Molecular plant, 14(8), 1362-1378.
- Shahan, R., Hsu, C. W., Nolan, T. M., Cole, B. J., Taylor, I. W., Greenstreet, L., … & Ohler, U. (2022). A single-cell Arabidopsis root atlas reveals developmental trajectories in wild-type and cell identity mutants. Developmental cell, 57(4), 543-560.
- Zhang, T. Q., Chen, Y., & Wang, J. W. (2021). A single-cell analysis of the Arabidopsis vegetative shoot apex. Developmental cell, 56(7), 1056-1074.
- Kim, J. Y., Symeonidi, E., Pang, T. Y., Denyer, T., Weidauer, D., Bezrutczyk, M., … & Frommer, W. B. (2021). Distinct identities of leaf phloem cells revealed by single cell transcriptomics. The Plant Cell, 33(3), 511-530.
- Liu, W., Zhang, Y., Fang, X., Tran, S., Zhai, N., Yang, Z., … & Xu, L. (2022). Transcriptional landscapes of de novo root regeneration from detached Arabidopsis leaves revealed by time-lapse and single-cell RNA sequencing analyses. Plant Communications, 3(4).
- Kang, M., Choi, Y., Kim, H., & Kim, S. G. (2022). Single‐cell RNA‐sequencing of Nicotiana attenuata corolla cells reveals the biosynthetic pathway of a floral scent. New Phytologist, 234(2), 527-544.
- Chen, H., Yin, X., Guo, L., Yao, J., Ding, Y., Xu, X., … & Fan, L. (2021). PlantscRNAdb: a database for plant single-cell RNA analysis. Molecular Plant, 14(6), 855-857.
- Xu, Z., Wang, Q., Zhu, X., Wang, G., Qin, Y., Ding, F., … & Jin, S. (2022). Plant Single Cell Transcriptome Hub (PsctH): an integrated online tool to explore the plant single‐cell transcriptome landscape. Plant Biotechnology Journal, 20(1), 10.
- Jin, J., Lu, P., Xu, Y., Tao, J., Li, Z., Wang, S., … & Cao, P. (2022). PCMDB: a curated and comprehensive resource of plant cell markers. Nucleic acids research, 50(D1), D1448-D1455.
- Jha, S. G., Borowsky, A. T., Cole, B. J., Fahlgren, N., Farmer, A., Huang, S. S. C., … & Rhee, S. Y. (2021). Vision, challenges and opportunities for a Plant Cell Atlas. Elife, 10, e66877.
______________________________________________
About the Author
Indrani Kakati is a postdoctoral researcher mother and a 2023 Plantae Fellow, wishing to connect with researchers around the globe and impart knowledge through communicating innovative research and wants to inspire and motivate young girls to take up science and never give up. You can find her on X/Twitter at @indranikb.