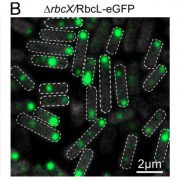
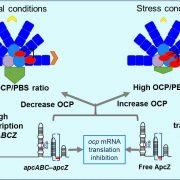
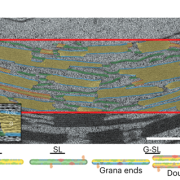
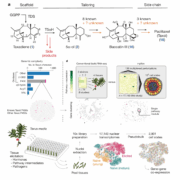
A CRISPRi library screen reveals growth-robustness tradeoffs in Synechocystis
Miao & Jahn et al. generated and screened a large CRISPRi-based library of Synechocystis in 11 different growth conditions to reveal growth-robustness tradeoffs and to discover genes with condition specific importance and additional functions.
https://doi.org/10.1093/plcell/koad208
By Rui Miao1, Michael Jahn1,2, and Elton P. Hudson1
1School of Engineering Sciences in Chemistry, Biotechnology and Health, Science for Life Laboratory, KTH – Royal Institute of Technology, Stockholm, Sweden.
2Max Planck Unit for the Science of Pathogens, Berlin, Germany.
Background: Cyanobacteria are photosynthetic microbes that play a crucial role in the global carbon cycle. They convert sunlight into chemical energy and produce oxygen as a byproduct. Extant cyanobacteria are also thought to share a common ancestor with the chloroplasts of plants and algae. However, nearly half of the genes in model cyanobacteria strains are not annotated with a function. This knowledge gap is a significant obstacle for fully understanding the physiology of photosynthesis, as well as for developing cyanobacteria into efficient cell factories for bioproduction.
Question: We aimed to map whether genes in cyanobactera are essential and contribute to cell fitness different growth conditions spanning a wide range of light regimes and carbon source availabilities.
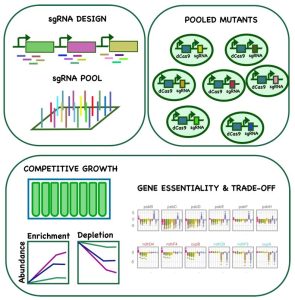 Findings: We screened a large gene repression library consisting of more than 20,000 individual mutants in 11 different conditions. We found that certain genes showed fitness benefits in one condition but were generally detrimental in others, including several genes that play critical roles in mixotrophy and photoheterotrophy, indicating their significance in redirecting metabolic flux. We also discovered genes with condition-specific importance and identified additional functions for central metabolic enzymes. We compiled the fitness data for all genes in all conditions in an interactive, open access web application. In addition, we used our extensive dataset in combination with machine learning to identify “design rules” for effective guide RNAs in the cyanobacterium Synechocystis.
Findings: We screened a large gene repression library consisting of more than 20,000 individual mutants in 11 different conditions. We found that certain genes showed fitness benefits in one condition but were generally detrimental in others, including several genes that play critical roles in mixotrophy and photoheterotrophy, indicating their significance in redirecting metabolic flux. We also discovered genes with condition-specific importance and identified additional functions for central metabolic enzymes. We compiled the fitness data for all genes in all conditions in an interactive, open access web application. In addition, we used our extensive dataset in combination with machine learning to identify “design rules” for effective guide RNAs in the cyanobacterium Synechocystis.
Next steps: Our results provide important insight to the cyanobacteria community, and to plant scientists looking for cyanobacterial homologues to chloroplast proteins or other examples of suboptimality in regulation of photoprotection. Future studies may focus on investigating the highlighted genes or regulatory pathways with individual mutants.
Rui Miao, Michael Jahn, Kiyan Shabestary, Gilles Peltier, Elton Paul Hudson. (2023). CRISPR interference screens reveal growth–robustness tradeoffs in Synechocystis sp. PCC 6803 across growth conditions. https://doi.org/10.1093/plcell/koad208