Prediction of conserved variable heat and cold stress response in maize using cis-regulatory information (Plant Cell)
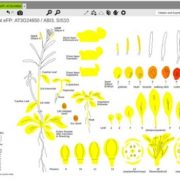
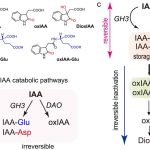
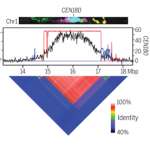
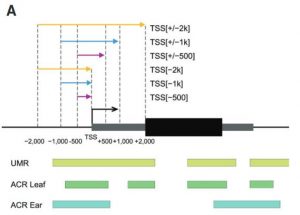 Several transcription factors (TFs) have been identified previously as responsive to cold and heat stresses in maize. Variation in TF binding sites (TFBS) determine changes in the expression of target genes in response to the aforementioned stresses. Traditionally, single varieties were used to study TFBS variation rather than different species which are, putatively, a source of cis-regulatory variation. To deepen this aspect, in this work Zhou et al. compared inbred and hybrid genotype responses after applying different heat/cold stresses. In addition, they developed machine learning models for predicting gene expression responses in new genotypes, highlighting the critical parameters for motif detection. To conclude, they point out that insertion/deletion (InDel) polymorphisms have a fundamental role in the cis-regulatory elements of the genes with differential responses to heat/cold stress. This work is a step further to study the link between the cis-regulatory variation, gene expression, and plant phenotypes. (Summary by Eva Maria Gomez Alvarez, @eva_ga96) Plant Cell 10.1093/plcell/koab267.
Several transcription factors (TFs) have been identified previously as responsive to cold and heat stresses in maize. Variation in TF binding sites (TFBS) determine changes in the expression of target genes in response to the aforementioned stresses. Traditionally, single varieties were used to study TFBS variation rather than different species which are, putatively, a source of cis-regulatory variation. To deepen this aspect, in this work Zhou et al. compared inbred and hybrid genotype responses after applying different heat/cold stresses. In addition, they developed machine learning models for predicting gene expression responses in new genotypes, highlighting the critical parameters for motif detection. To conclude, they point out that insertion/deletion (InDel) polymorphisms have a fundamental role in the cis-regulatory elements of the genes with differential responses to heat/cold stress. This work is a step further to study the link between the cis-regulatory variation, gene expression, and plant phenotypes. (Summary by Eva Maria Gomez Alvarez, @eva_ga96) Plant Cell 10.1093/plcell/koab267.