Plant Science Research Weekly: January 17
Guest Editor :
Facundo Romani
I am in an Argentinean PhD student at Universidad Nacional del Litoral (Santa Fe, Argentina) and current ASPB Plantae Fellow. I am about to finish my PhD program, during past years I worked with Javier Moreno as supervisor and spent a long stay at Monash University (Australia) in John Bowman’s lab. My research focus is on macroevolution of transcriptional regulation in plant development and environmental responses. I am interested in evo-devo and Marchantia polymorpha. Twitter: @facundoromani
R-Loop Mediated trans Action of the APOLO Long Noncoding RNA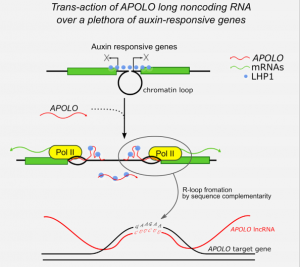
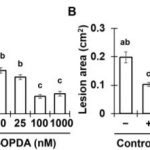
It was shown that the Arabidopsis intergenic long noncoding RNA (LncRNA) APOLO (AUXIN-REGULATED PROMOTER LOOP) modulates in cis its neighbor gene PID forming a chromatin loop. However, it remained unknown whether it can act also in trans modulating the expression of other genes. Now, Ariel et al. performed chromatin isolation by APOLO-RNA purification followed by DNA-seq (ChIRP-seq) and RNA-seq of overexpressing APOLO lines in order to identify potential targets in the Arabidopsis genome. They found a group of “bona fide” targets also related to auxin response. These target loci are also enriched in the binding of the Polycomb Repressive Complex 1 (PRC1) component LHP1 and the formation of DNA-RNA duplex (R-loops). They propose a model where APOLO is mediating the formation of R-loops and decoying the PRC1 complex over target loci to modulate gene expression. In order to demonstrate that, they performed DRIP-qPCR and developed a novel method named RNA isolation by DNA purification (RIDP) to purify a specific DNA locus to further isolate RNA and quantify R-loop dependent APOLO presence using RNAseH. This paper shows that lncRNA can shape the three-dimensional organization of genetic information and are a promising source of distal transcriptional regulators in plants. (Summary by Facundo Romani) Molecular Cell 10.1016/j.molcel.2019.12.015
Special Issue: New perspectives on crassulacean acid metabolism biology
 Crassulacean acid metabolism (CAM) is a water-conserving strategy in which stomata open at night and carbon is stored until daytime photosynthesis provides the energy to fix it. This special issue of the Journal of Experimental Botany, edited by Hultine, Cushman, and Williams, brings together a set of papers that explore new insights into the evolution and variation of this pathway. As the editors observe, this is complex trait is ripe for comparative analysis, as it has, “>60 independent evolutionary origins, and occurrence found in >38 families encompassing >400 genera of vascular plant species.” Furthermore, there are numerous plants that exhibit facultative CAM or variations on CAM. The issue also includes discussions about potential agricultural opportunities that can come from exploiting the water-saving properties of CAM (as examples, familiar CAM plants include pineapple, agave and vanilla). (Summary by Mary Williams) J. Exp. Bot. 70: Issue 22.
Crassulacean acid metabolism (CAM) is a water-conserving strategy in which stomata open at night and carbon is stored until daytime photosynthesis provides the energy to fix it. This special issue of the Journal of Experimental Botany, edited by Hultine, Cushman, and Williams, brings together a set of papers that explore new insights into the evolution and variation of this pathway. As the editors observe, this is complex trait is ripe for comparative analysis, as it has, “>60 independent evolutionary origins, and occurrence found in >38 families encompassing >400 genera of vascular plant species.” Furthermore, there are numerous plants that exhibit facultative CAM or variations on CAM. The issue also includes discussions about potential agricultural opportunities that can come from exploiting the water-saving properties of CAM (as examples, familiar CAM plants include pineapple, agave and vanilla). (Summary by Mary Williams) J. Exp. Bot. 70: Issue 22.
The γ-tubulin complex protein GCP6 in crucial for spindle morphogenesis but not essential for microtubule reorganization in Arabidopsis (OA)
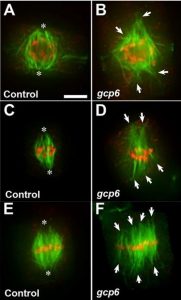
During cell division, cells require to form mitotic spindle and these mitotic spindles are basically newly formed microtubules. The formation of new microtubules depends on γ-tubulin. Along with 5 GPCs (γ-tubulin protein complex), γ-tubulin form γTuRC (γ-tubulin ring complex) and it facilitates the microtubule nucleation. Interestingly, in plant cells, microtubules are nucleated and organized without centrosome. In this article, Miao et al. investigated whether γTuRC is the only functional form of γ-tubulin for microtubule nucleation or another nucleation mechanism that exists in the plant cell. To test this idea, they have used the GPC6, which plays a major role as γTuRC protein in animal and fungal system, mutant (gpc6) in Arabidopsis thaliana and studied the mitotic division in detail. Surprisingly, they have found that although gpc6 mutant has a defect in γ-tubulin localization in microtubule array during mitotic division, these cells successfully complete cell division. This study indicates the major role of γTuRC for microtubule organization and provides insight about γTuRC-independent microtubule organization during mitotic division as well. (Summary by Arif Ashraf) PNAS 10.1073/pnas.1912240116
Review: Matrix redox physiology governs the regulation of plant mitochondrial metabolism through post-translational protein modifications
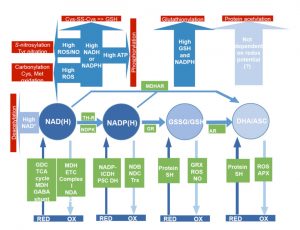
Mitochondrial metabolism provides ATP and reducing power to drive myriad reactions in the plant cell, and is constantly being fine-tuned in response to environment and demands. Post-translational modifications (PTMs) of proteins, including redox reactions of methionine and cysteine and carbamylation of lysine, arginine and proline, are crucial for this rapid, reversible responsiveness. The rate of mitochondrial electron transport affects the redox status of individual components, which can lead directly to post-translational modifications, hence alter metabolic flux, as reviewed by Møller et al. The authors discuss the reactions that determine the redox status of molecules including NAD(P)H/NAD(P)+, ascorbate, glutathione and thioredoxin, and how these molecules in turn affect PTMs and protein activities. They provide examples of adaptations and responses that occur during transitions such as light fluctuations and seed germination. (Summary by Mary Williams) Plant Cell 10.1105/tpc.19.00535
Direct conversion of carlactonoic acid to orobanchol by cytochrome P450 CYP722C in strigolactone biosynthesis
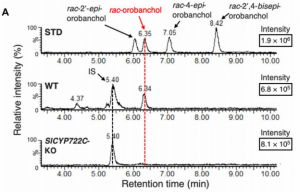
Strigolactones (SL) are a compounds that play important roles as phytohormones and as a rhizosphere signaling. Despite the great advances in their research that occurred recently, their biosynthetic pathway is still not well understood. Until now, the pathway leading to the precursor carlactonic acid (CLA) was described, where MAX1 is responsible for the last step. In this work, Takatoshi et al. identified a previously uncharacterized cytochrome P450 enzyme (CYP722C) involved in the direct biosynthesis of the canonical SL orobanchol and solanacol. This enzyme can convert CLA in orobanchol without the intermediate 4-Deoxy orobanchol as some MAX1 homologs do. They support their findings using in vitro enzyme assays and also produced a CRISPR knockout in tomato. Mutant plants did not present phenotypic defects in plant development and probably CYP722C does not explain most of SL biosynthesis. Despite that, authors provide evidence of the relevance of these compounds in rhizosphere signaling, showing impaired parasitic seed germination that could be of biotechnological interest. This article provides advances about the biosynthesis of this complex array of phytohormones, lots of exciting questions still awaits to be answered. (Summary by Facundo Romani) Science Advances 10.1126/sciadv.aax9067
Metabolite regulatory interactions control plant respiratory metabolism via Target of Rapamycin (TOR) kinase activation
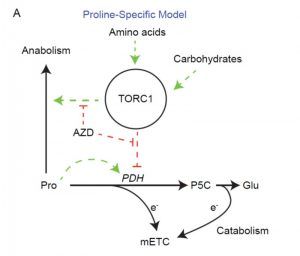
The rate of mitochondrial respiration is highly dynamic and responds to numerous endogenous and exogenous factors. Here, O’Leary et al. have measured mitochondrial respiration (through O2 production) in response to various metabolites in leaf discs over several hours, to address not only post-translational controls but also de novo transcriptional/translational effects. They found that, as expected, O2 production increases in response to carbohydrates (e.g., sucrose, fructose), but the strongest stimulation occurrs in response to the amino acids proline and alanine. This stimulation effect is diminished by inhibition of TOR kinase, a central hub integrating information about nutritional and environmental status. These results show that the effects of amino acids on O2 production are mediated by TOR. The authors propose a model in which the presence of amino acids signals nutrient abundance, via the TOR kinase pathway, influencing respiratory activity and thus plant metabolic rate. (Summary by Mary Williams) Plant Cell 10.1105/tpc.19.00157
Pheophorbide a may regulate jasmonate signaling during dark-induced senescence
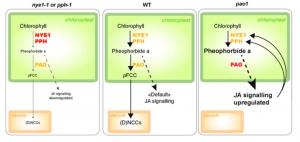
During leaf senescence, nitrogen-rich chlorophyll is broken down through a regulated process so that nitrogen-containing compounds can be reassimilated into the plant body. Chlorophyll catabolites are sequestered in the vacuole as linear tetrapyrroles known as phyllobilins, produced through the action of the porphyrin ring-opening activity of PHEOPHORBIDE A OXYGENASE (PAO); this is known as the PAO/phyllobilin pathway. Starting with loss-of-function mutants of PAO and two other chlorophyll-catabolic enzymes, Aubry et al. carried out a transcriptomic and metabolomic study during leaf senescence. The pao1 mutant accumulates pheophorbide a and shows a dramatically altered pattern of gene expression for chlorophyll degradation genes as well as for genes involved in jasmonic acid (JA) signaling and an increase in JA levels; JA is a known inducer of senescence. The authors propose “a retrograde signaling function for the PAO/phyllobilin pathway that uses JA-signaling to coordinate chloroplasts and the nucleus during dark-induced senescence.” (Summary by Mary Williams) Plant Physiol. 10.1104/pp.19.01115
Review: The impact of synthetic biology for future agriculture and nutrition
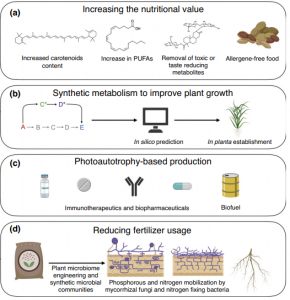 The synthetic biology field is going to be important for the decade we began in order to face climate challenges, including food security. However, plant synthetic biology lags behind bacterial and other eukaryotic systems. Roell and Zurbriggen summarize in this review, many of the projects that are currently taking place in this multidisciplinary subfield. They highlight how scientists around the world are engineering plants to improve nutrition quality removing undesirable metabolites and increasing beneficial ones. Plants can also help in the massive production of valuable biopharmaceutical and immunotherapeutic products thanks to their scalability. Scientists are also working on the improvement of plant autotrophic metabolism using synthetic biology. The reduction in fertilizer use is one of the main topics in the field too, that includes the use of synthetic bacterial communities, plant nitrogenases, and the establishment of the rhizobium-legume symbiosis. Decades ago, many promises were made about plant genetic engineering, some of them are coming on age in spite of limitations in the regulatory framework and will attract great attention in the future. (Summary by Facundo Romani) Curr. Opin. Biotech. 10.1016/j.copbio.2019.10.004
The synthetic biology field is going to be important for the decade we began in order to face climate challenges, including food security. However, plant synthetic biology lags behind bacterial and other eukaryotic systems. Roell and Zurbriggen summarize in this review, many of the projects that are currently taking place in this multidisciplinary subfield. They highlight how scientists around the world are engineering plants to improve nutrition quality removing undesirable metabolites and increasing beneficial ones. Plants can also help in the massive production of valuable biopharmaceutical and immunotherapeutic products thanks to their scalability. Scientists are also working on the improvement of plant autotrophic metabolism using synthetic biology. The reduction in fertilizer use is one of the main topics in the field too, that includes the use of synthetic bacterial communities, plant nitrogenases, and the establishment of the rhizobium-legume symbiosis. Decades ago, many promises were made about plant genetic engineering, some of them are coming on age in spite of limitations in the regulatory framework and will attract great attention in the future. (Summary by Facundo Romani) Curr. Opin. Biotech. 10.1016/j.copbio.2019.10.004