Plant Science Research Weekly: December 27
Special Current Opinion in Biotechnology issue Issue: Plant Biotechnology
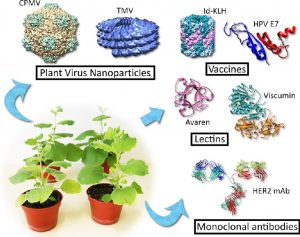 Since most of us will have a few days off this coming week as we welcome in 2020, I’d like to highlight some of the engrossing reviews in this special issue of Current Opinion in Biotechnology, edited by Ralf Reski, Gary Foster & Ed Rybicki. Several of the articles focus on Molecular Pharming, or the production of high-value medicinal products in plants. Such efforts vaulted into the headlines with the use of ZMapp (a cocktail of plant-made monoclonal antibodies) during the 2014–2016 Ebola virus outbreak in West Africa. Some of the reviews in this Special Issue focus on products (Molecular farming for therapies and vaccines in Africa; Cancer biologics made in plants), others on strategies for increasing the efficiency or versatility of plants as pharmaceutical producers (Proteases of Nicotiana benthamiana: an emerging battle for molecular farming; Optimizing product quality in molecular farming; Innovation in plant-based transient protein expression for infectious disease prevention and preparedness), and others on the value or optimization of specific types of plants (Green algal hydrocarbon metabolism is an exceptional source of sustainable chemicals; Mosses in biotechnology). There are also useful reviews of progress and policies concerning gene-edited plants. (Summary by Mary Williams) Curr. Opin. Biotech. Feb. 2020.
Since most of us will have a few days off this coming week as we welcome in 2020, I’d like to highlight some of the engrossing reviews in this special issue of Current Opinion in Biotechnology, edited by Ralf Reski, Gary Foster & Ed Rybicki. Several of the articles focus on Molecular Pharming, or the production of high-value medicinal products in plants. Such efforts vaulted into the headlines with the use of ZMapp (a cocktail of plant-made monoclonal antibodies) during the 2014–2016 Ebola virus outbreak in West Africa. Some of the reviews in this Special Issue focus on products (Molecular farming for therapies and vaccines in Africa; Cancer biologics made in plants), others on strategies for increasing the efficiency or versatility of plants as pharmaceutical producers (Proteases of Nicotiana benthamiana: an emerging battle for molecular farming; Optimizing product quality in molecular farming; Innovation in plant-based transient protein expression for infectious disease prevention and preparedness), and others on the value or optimization of specific types of plants (Green algal hydrocarbon metabolism is an exceptional source of sustainable chemicals; Mosses in biotechnology). There are also useful reviews of progress and policies concerning gene-edited plants. (Summary by Mary Williams) Curr. Opin. Biotech. Feb. 2020.
Review: Molecular bases of responses to abiotic stress in trees
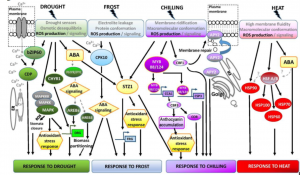 Much of our understanding of abiotic stress response comes from studies on short-lived annual plants, for good reasons: they are small and easy to study, their short-generation time makes them amenable to genetic studies, and most of our food comes from annual plants. Trees are more difficult to study, but no less important in their roles as ecosystem service providers, sources of wood and fibers, and major global carbon sinks. As large, long-lived organisms, trees face additional abiotic challenges, such as long-distance water transport and coping with widely varying seasonal temperatures. Worryingly, trees are already showing their vulnerability to climate change. Estravis-Barcala et al. look the molecular bases of tree responses to abiotic stress, including short-term acclimation responses and longer-term adaptation responses, pulling together new studies from a range of tree species. They also summarize efforts to identify the genetic basis for adaptation to local environments through genome scanning and the identification of selection signatures, which have pinpointed selected dehydrins and histones as contributors. The review also usefully highlights some of the key knowledge gaps that remain to be addressed. (Summary by Mary Williams) J. Exp. Bot. 10.1093/jxb/erz532
Much of our understanding of abiotic stress response comes from studies on short-lived annual plants, for good reasons: they are small and easy to study, their short-generation time makes them amenable to genetic studies, and most of our food comes from annual plants. Trees are more difficult to study, but no less important in their roles as ecosystem service providers, sources of wood and fibers, and major global carbon sinks. As large, long-lived organisms, trees face additional abiotic challenges, such as long-distance water transport and coping with widely varying seasonal temperatures. Worryingly, trees are already showing their vulnerability to climate change. Estravis-Barcala et al. look the molecular bases of tree responses to abiotic stress, including short-term acclimation responses and longer-term adaptation responses, pulling together new studies from a range of tree species. They also summarize efforts to identify the genetic basis for adaptation to local environments through genome scanning and the identification of selection signatures, which have pinpointed selected dehydrins and histones as contributors. The review also usefully highlights some of the key knowledge gaps that remain to be addressed. (Summary by Mary Williams) J. Exp. Bot. 10.1093/jxb/erz532
Review: Surface sensor systems in plant immunity
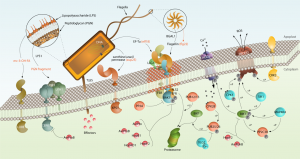 The first line of defense is detection. Plants have numerous cell-surface receptor proteins (Pattern Recognition Receptors, PRRs) that recognize potentially harmful pathogens as well as endogenous molecules that suggest damage, known as Damage Associated Molecular Patterns or DAMPS and phytocytokines (not to be confused with cytokinins!). If you’re not working in this area, it can come across as a bit of an “alphabet soup”, particularly as the PRRs are found in large gene families and are frequently genus-specific due to “substantial evolutionary dynamics.” This review by Albert et al. does a nice job of putting all of these abbreviations into context to help you sort out your LRR-RKs from your LRR-RPs from your LysM-RK/RPs. The signaling pathways downstream of the PRRs are also covered, as are the effectors produced by pathogens that target PRRs. (Summary by Mary Williams) Plant Physiol. 10.1104/pp.19.01299
The first line of defense is detection. Plants have numerous cell-surface receptor proteins (Pattern Recognition Receptors, PRRs) that recognize potentially harmful pathogens as well as endogenous molecules that suggest damage, known as Damage Associated Molecular Patterns or DAMPS and phytocytokines (not to be confused with cytokinins!). If you’re not working in this area, it can come across as a bit of an “alphabet soup”, particularly as the PRRs are found in large gene families and are frequently genus-specific due to “substantial evolutionary dynamics.” This review by Albert et al. does a nice job of putting all of these abbreviations into context to help you sort out your LRR-RKs from your LRR-RPs from your LysM-RK/RPs. The signaling pathways downstream of the PRRs are also covered, as are the effectors produced by pathogens that target PRRs. (Summary by Mary Williams) Plant Physiol. 10.1104/pp.19.01299
Control of nitrogen fixation in bacteria that associate with cereals
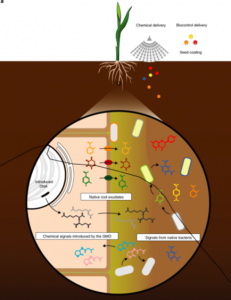 Legumes benefit from mutualistic symbioses with Rhizobia bacteria, which are induced to fix nitrogen when inside of the nodules produced by the plant. Cereals also associate with Rhizobia, but without nodules they don’t fix nitrogen. Ryu et al. set out to engineer inducible nitrogenase activity in free-living Rhizobia and other bacteria that associate with cereals. They introduced natural and engineered nitrogen fixing (nif) gene clusters from several different bacteria sources into cereal-associating bacteria. Some operons were engineered to remove inhibition by ammonium (the end-product of nitrogenase), and some were engineered to be induced by synthetic genetically encoded sensors. In the presense of plants that activate these sensors, nif transcription is induced. Some of these studies led to bacteria that are able to fix nitrogen in the presence of plants, a first step in building strains that can efficiently deliver fixed nitrogen to cereals. (Summary by Mary Williams) Nature Microbiol. 10.1038/s41564-019-0631-2
Legumes benefit from mutualistic symbioses with Rhizobia bacteria, which are induced to fix nitrogen when inside of the nodules produced by the plant. Cereals also associate with Rhizobia, but without nodules they don’t fix nitrogen. Ryu et al. set out to engineer inducible nitrogenase activity in free-living Rhizobia and other bacteria that associate with cereals. They introduced natural and engineered nitrogen fixing (nif) gene clusters from several different bacteria sources into cereal-associating bacteria. Some operons were engineered to remove inhibition by ammonium (the end-product of nitrogenase), and some were engineered to be induced by synthetic genetically encoded sensors. In the presense of plants that activate these sensors, nif transcription is induced. Some of these studies led to bacteria that are able to fix nitrogen in the presence of plants, a first step in building strains that can efficiently deliver fixed nitrogen to cereals. (Summary by Mary Williams) Nature Microbiol. 10.1038/s41564-019-0631-2
Cleavage of a pathogen apoplastic protein by plant subtilases activates immunity
 Plant-pathogen interactions are shaped by a dynamic signaling crosstalk that often leads to an arms-race between plants and pathogens. The initial pathogenic invasion starts in the apoplast, which serves as a major battlefield. This extracellular space is a harsh environment enriched with hydrolytic enzymes such as proteases. Previously, an extracellular protease has been identified as a pathogen-induced protein, but its function has been elusive. Here, Wang et al. show a classic example of the co-evolving defense mechanism in a plant-pathogen interaction. The authors developed a pipeline to identify cysteine-rich secreted peptides from Phytophthora infestans. They identified PC2, which has broad sequence conservation in oomycetes but shows functional divergence. PC2 activity depends on cleavage by a plant apoplastic serine protease. Upon PC2 cleavage, plant immunity is activated, leading to cell death. To evade this plant surveillance system, P. infestans has evolved protease inhibitors that block PC2 cleavage. (Summary by Amey Redkar) bioRxiv. 10.1101/2019.12.16.878272v1
Plant-pathogen interactions are shaped by a dynamic signaling crosstalk that often leads to an arms-race between plants and pathogens. The initial pathogenic invasion starts in the apoplast, which serves as a major battlefield. This extracellular space is a harsh environment enriched with hydrolytic enzymes such as proteases. Previously, an extracellular protease has been identified as a pathogen-induced protein, but its function has been elusive. Here, Wang et al. show a classic example of the co-evolving defense mechanism in a plant-pathogen interaction. The authors developed a pipeline to identify cysteine-rich secreted peptides from Phytophthora infestans. They identified PC2, which has broad sequence conservation in oomycetes but shows functional divergence. PC2 activity depends on cleavage by a plant apoplastic serine protease. Upon PC2 cleavage, plant immunity is activated, leading to cell death. To evade this plant surveillance system, P. infestans has evolved protease inhibitors that block PC2 cleavage. (Summary by Amey Redkar) bioRxiv. 10.1101/2019.12.16.878272v1
Successive passaging of plant associated microbiome reveals robust habitat and host genotype-dependent selection
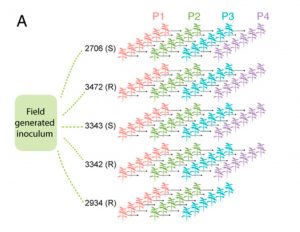 Study of the plant microbiome can contribute to sustainable agricultural systems through improvements to plant health and nutrition. One goal has been to select and design microbiome communities for specific functions. In a recent article, Morella et al. have used an experimental evolution approach to understand the robustness of the plant microbiome after colonization of the host. They inoculated five different genotypes of tomato with a diverse set of microbes, which were serially passaged four times from 45-day old plants. The phyllosphere bacterial and fungal communities were sequenced and analyzed after each passage, which revealed a loss of diversity and a loss of genotype effect. At the end of the first passage, 92% of initial inoculum operational taxonomic units (OTUs) were present, and the number reduced to 29% at the end of the fourth passage. Interestingly, the bacterial community from the end of the experiment was robust to invasion by the starting bacterial community. This indicates that efforts to support plant growth by inoculating them with a beneficial microbiome could be successful. (Summary by Mugdha Sabale) Proc. Natl. Acad. Sci. USA
Study of the plant microbiome can contribute to sustainable agricultural systems through improvements to plant health and nutrition. One goal has been to select and design microbiome communities for specific functions. In a recent article, Morella et al. have used an experimental evolution approach to understand the robustness of the plant microbiome after colonization of the host. They inoculated five different genotypes of tomato with a diverse set of microbes, which were serially passaged four times from 45-day old plants. The phyllosphere bacterial and fungal communities were sequenced and analyzed after each passage, which revealed a loss of diversity and a loss of genotype effect. At the end of the first passage, 92% of initial inoculum operational taxonomic units (OTUs) were present, and the number reduced to 29% at the end of the fourth passage. Interestingly, the bacterial community from the end of the experiment was robust to invasion by the starting bacterial community. This indicates that efforts to support plant growth by inoculating them with a beneficial microbiome could be successful. (Summary by Mugdha Sabale) Proc. Natl. Acad. Sci. USA
An artificial metalloenzyme biosensor can detect ethylene gas in fruits and Arabidopsis leaves
 Biosensors have provided abundant information about the distribution of target enzymes, for example auxin, revealing much about their roles in plant physiology and development. Ethylene is a tiny (C2H4) gaseous hormone that hasn’t been very amenable to the development of biosensors. Here, Vong et al. describe the development of an albumin-based artificial metalloenzyme (ArM) that binds a ruthenium metal complex along with a fluorescence molecule and a quencher. In the absence of ethylene, the quencher prevents fluorescence, but in ethylene’s presence the quencher is released and a fluorescence signal formed. This ArM ethylene probe (AEP) fluoresces in the presence of ethylene, as demonstrated in various tissues and in response to pathogens. (Summary by Mary Williams) Nature Comms. 10.1038/s41467-019-13758-2
Biosensors have provided abundant information about the distribution of target enzymes, for example auxin, revealing much about their roles in plant physiology and development. Ethylene is a tiny (C2H4) gaseous hormone that hasn’t been very amenable to the development of biosensors. Here, Vong et al. describe the development of an albumin-based artificial metalloenzyme (ArM) that binds a ruthenium metal complex along with a fluorescence molecule and a quencher. In the absence of ethylene, the quencher prevents fluorescence, but in ethylene’s presence the quencher is released and a fluorescence signal formed. This ArM ethylene probe (AEP) fluoresces in the presence of ethylene, as demonstrated in various tissues and in response to pathogens. (Summary by Mary Williams) Nature Comms. 10.1038/s41467-019-13758-2
A practical guide for fluorophore selection for FRET experiments in plants
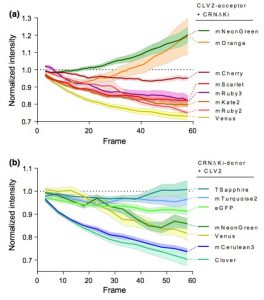 Protein-protein interactions modulate the activities of many proteins, i.e., the signaling specificity of Receptor-like kinases (RLK). Förster Resonance Energy Transfer (FRET) allows the study of the dynamics of protein-protein interactions in live plants; this technique is based on energy transfer from a fluorescent donor to an acceptor fluorophore. There are many fluorophore pairs available for FRET experiments, but most of them have been characterized in vitro or in mammalian cell systems. In this study, Denay and collaborators tested the performance of 12 fluorophore pairs in a plant cell system. They focused on membrane proteins; for this, they chose the CORYNE-CLAVATA2 (CRN-CLV2) dimer. Protein localization analysis allowed the study of the fluorophore effect on protein folding and function since a correct CRN/CLV2 interaction is necessary for a correct export from the endoplasmic reticulum to the plasma membrane. Other analyses on the fluorophores included sensitivity to photobleaching and noise-to-signal ratio in the cellular context. Measurement of FRET efficiency confirmed eGFP-mCherry as a good FRET pair, as well as other combinations with similar performance, i.e., Venus-mKate2, Venus-mRuby3, mCerulean3-mNeonGreen, and mTurquoise2-Venus. Furthermore, the authors identified combinations of three different fluorophores that would allow the study of interactions between three proteins, protein complexes, or the measurement of competitive interactions. (Summary by Humberto Herrera-Ubaldo) Plant Direct. 10.1002/pld3.189
Protein-protein interactions modulate the activities of many proteins, i.e., the signaling specificity of Receptor-like kinases (RLK). Förster Resonance Energy Transfer (FRET) allows the study of the dynamics of protein-protein interactions in live plants; this technique is based on energy transfer from a fluorescent donor to an acceptor fluorophore. There are many fluorophore pairs available for FRET experiments, but most of them have been characterized in vitro or in mammalian cell systems. In this study, Denay and collaborators tested the performance of 12 fluorophore pairs in a plant cell system. They focused on membrane proteins; for this, they chose the CORYNE-CLAVATA2 (CRN-CLV2) dimer. Protein localization analysis allowed the study of the fluorophore effect on protein folding and function since a correct CRN/CLV2 interaction is necessary for a correct export from the endoplasmic reticulum to the plasma membrane. Other analyses on the fluorophores included sensitivity to photobleaching and noise-to-signal ratio in the cellular context. Measurement of FRET efficiency confirmed eGFP-mCherry as a good FRET pair, as well as other combinations with similar performance, i.e., Venus-mKate2, Venus-mRuby3, mCerulean3-mNeonGreen, and mTurquoise2-Venus. Furthermore, the authors identified combinations of three different fluorophores that would allow the study of interactions between three proteins, protein complexes, or the measurement of competitive interactions. (Summary by Humberto Herrera-Ubaldo) Plant Direct. 10.1002/pld3.189
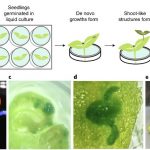
Plant gene editing through de novo induction of meristems ($)
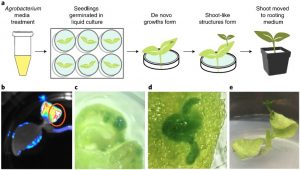 A fast method of gene editing using Agrobacterium was developed to deliver combinations of the developmental regulators including WUSCHEL and SHOOT MERISTEMLESS along with gene-editing reagents. The expression of specific developmental regulators led to the induction of meristems and expression of the regulators was monitored by the tagged Luciferase marker expression. The shoot-like tissue was subsequently transferred to rooting medium to regenerate whole, fertile plants. To monitor gene editing, guide RNA targeting phytoene desaturase genes was also co-expressed. Successful genome editing was performed in the induced meristems of cultured tobacco. In a further study, the Agrobacteria were applied to other soil-grown dicots (potato, grape) from which the meristematic tissue had been removed, and some of the resulting growth had evidence of gene editing. Overall this new approach will enhance genome editing in multiple dicots by alleviating the issues associated with tissue culture. (Summary by Suresh Damodaran). Nature Biotech. 10.1038/s41587-019-0337-2
A fast method of gene editing using Agrobacterium was developed to deliver combinations of the developmental regulators including WUSCHEL and SHOOT MERISTEMLESS along with gene-editing reagents. The expression of specific developmental regulators led to the induction of meristems and expression of the regulators was monitored by the tagged Luciferase marker expression. The shoot-like tissue was subsequently transferred to rooting medium to regenerate whole, fertile plants. To monitor gene editing, guide RNA targeting phytoene desaturase genes was also co-expressed. Successful genome editing was performed in the induced meristems of cultured tobacco. In a further study, the Agrobacteria were applied to other soil-grown dicots (potato, grape) from which the meristematic tissue had been removed, and some of the resulting growth had evidence of gene editing. Overall this new approach will enhance genome editing in multiple dicots by alleviating the issues associated with tissue culture. (Summary by Suresh Damodaran). Nature Biotech. 10.1038/s41587-019-0337-2
Rapid customization of Solanaceae fruit crops for urban agriculture ($)
 Numerous genes have been identified that modify shoot architecture, which has allowed breeding of varieties for specific purposes and environments. Here, Kwon et al. describe how they have used gene editing to modify several of these genes to produce tomatoes and groundcherries that are compact and rapid cycling, so are optimized for restricted growth environments such as indoor farming systems and possibly even during space travel. (Summary by Mary Williams) Nature Biotech. 10.1038/s41587-019-0361-2
Numerous genes have been identified that modify shoot architecture, which has allowed breeding of varieties for specific purposes and environments. Here, Kwon et al. describe how they have used gene editing to modify several of these genes to produce tomatoes and groundcherries that are compact and rapid cycling, so are optimized for restricted growth environments such as indoor farming systems and possibly even during space travel. (Summary by Mary Williams) Nature Biotech. 10.1038/s41587-019-0361-2