Stiffening stems: identification of the stiff1 gene involved in maize stalk strength
Stalk lodging, a structural failure in which crop stalks break prior to harvest, can result in high grain moisture, reduce grain quality, and cause harvesting difficulties. For the popular high-yielding cereal maize (Zea mays), lodging may result in global annual yield reductions of approximately 5-20% (Flint-Garcia et al., 2003). Developing maize lines with improved stalk strength is therefore a major breeding interest. Although stalk strength has been shown to be controlled by numerous quantitative trait loci (QTLs), the underlying genetic factors and their molecular functions remain largely unknown. Recent elegant work from Zhang and colleagues (2019) mapped and characterised the gene stiff1, whose repression causes improved stalk strength.
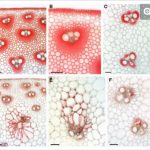
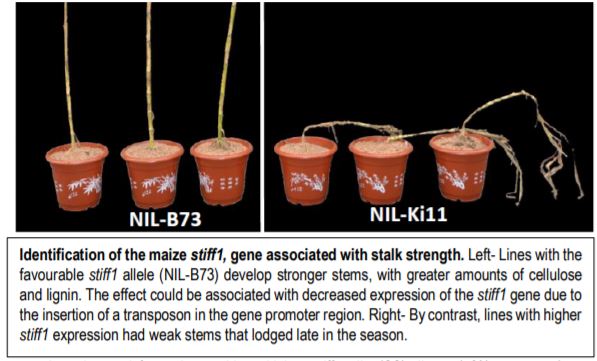
The stiff1 QTL was isolated as a major determinant of stalk strength variation from analysis of recombinant inbred lines produced by crossing the stiff-stalk line B73 with the non-stiff-stalk line Ki11. Subsequent comparison of two near isogenic lines (NILs) homozygous for either the B73 or Ki11 stiff1 allele revealed that NIL-B73 plants had significantly stronger stalks, increased plant height and improved stalk diameter traits, as well as increased cellulose and lignin. By contrast, NIL-Ki11 plants had weak stalks and lodged in the late season.
 Exhaustive fine mapping narrowed the stiff1 gene location to a genomic region containing just two gene candidates, of which, only one – encoding an F-box domain protein- was expressed in the stalk. Analysis of sequence variations in the stiff1 candidate gene region between the strong-stemmed B73 and Ki11 revealed multiple insertions, deletions, and single-nucleotide polymorphisms. By performing association mapping of these polymorphisms across 256 maize accession, two conspicuous insertions in the gene promoter region were found to have a probable causal role in the stiff1 phenotype: a 578 bp insertion and a 27.2-kb insertion consisting of four Ty1/Copia transposable elements. Further luciferase transient assays revealed that only the 27.2-kb insertion was causative to stiff stalk. Transposons make up the majority of the maize genome and have been linked to important stages of maize domestication, including evolution from seed shattering to non-shattering and from plants with multiple small ears to a single large ear. This work suggests a possible role for transposons in further maize improvement.
Exhaustive fine mapping narrowed the stiff1 gene location to a genomic region containing just two gene candidates, of which, only one – encoding an F-box domain protein- was expressed in the stalk. Analysis of sequence variations in the stiff1 candidate gene region between the strong-stemmed B73 and Ki11 revealed multiple insertions, deletions, and single-nucleotide polymorphisms. By performing association mapping of these polymorphisms across 256 maize accession, two conspicuous insertions in the gene promoter region were found to have a probable causal role in the stiff1 phenotype: a 578 bp insertion and a 27.2-kb insertion consisting of four Ty1/Copia transposable elements. Further luciferase transient assays revealed that only the 27.2-kb insertion was causative to stiff stalk. Transposons make up the majority of the maize genome and have been linked to important stages of maize domestication, including evolution from seed shattering to non-shattering and from plants with multiple small ears to a single large ear. This work suggests a possible role for transposons in further maize improvement.
The function of stiff1 in stem stability was supported by the creation and analysis of ubiquitin promoter-driven stiff1 overexpression lines (Ovstiff1), and a CRISPR/Cas9 stiff1 disrupted line (Cstiff1). While 3 out of 5 independently obtained Ovstiff1 lines had thinner, softer stalks, reduced stiffness, and lodged in the late season, Cstiff1 had significantly increased stalk strength, indicating a negative relationship between the newly assigned gene and stiffness. Similarly, the stronger NIL-B73 lines were shown to have reduced accumulation of stiff1 mRNA relative to NIL-Ki11 plants, an effect that could be linked to disruption of an upstream mini-enhancer element by the large transposon insertion. Both NIL-B73 and Cstiff1 line exhibited significantly increased cellulose and lignin levels in the stalk, and showed thicker-walled schlerenchyma cells, compared with Ki11 and Ovstiff. Stiff1 was shown to be expressed mostly in the vascular bundles, with the protein localised to the cytoplasm and nucleus. RNA-seq analysis between Cstiff1 and control plants suggested that stiff1 regulates factors involved in several hormone signalling pathways.
Analysis of the global natural population of 265 maize inbred lines revealed an increased presence of the favourable B73-type stiff1 allele in lines previously categorised as stiff-stalk (SS) lines (50% presence), as opposed to those considered nonstiff-stalk or tropical-subtropical (Flint-Garcia et al., 2005). The incredibly low prevalence of the favourable allele in groups outside of the SS lines suggests that precise gene editing of stiff1 in non-SS plants may provide a new and rapid method for greatly improving maize stalk strength on a large scale.
References:
Flint-Garcia, S.A., Jampatong, C., Darrah, L.L., and McMullen, M.D. (2003a). Quantitative trait locus analysis of stalk strength in four maize populations. Crop Sci 43, 13-22.
Flint-Garcia, S.A., Thuillet, A.C., Yu, J., Pressoir, G., Romero, S.M., Mitchell, S.E., Doebley, J., Kresovich, S., Goodman, M.M., and Buckler, E.S. (2005). Maize association population: a high-resolution platform for quantitative trait locus dissection. The Plant Journal 44, 1054-1064.
Zhang, Z., Zhang, X., Lin, Z., Wang, J., Liu, H., Zhou, L., Zhong, S., Li, Y., Zhu, C., Lai, J., Li, X., Yu, J., Lin, Z. (2019). A large transposon insertion in the stiff1 promoter controls stalk strength in maize. The Plant Cell. https://doi.org/10.1105/tpc.19.00486