Patterns and consequences of subgenome differentiation provide insights into the nature of paleopolyploidy in plants
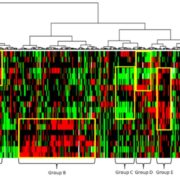
 Many plants are polyploid, meaning that instead of the normal, diploid “2n” complement of chromosomes (one from each parent), they have more than 2n. Following whole-genome duplication, redundancy can allow the duplicated regions to diverge or become silenced. In some cases, silencing occurs preferentially in one of the two parental genomes. By comparing the genomes of maize and soybean to their diploid relatives, Zhao et al. compared the consequences of (ancient) paleopolyploidy events in two species. They found that in maize, “one subgenome has been subject to relaxed selection, lower levels of gene expression, higher rates of transposable element accumulation and more small interfering RNAs and DNA methylation around genes, and higher rates of gene loss,” whereas although soybean does not exhibit subgenome dominance, “members of gene pairs do become differentiated, and that differentiation is associated with several of the same markers of differentiation that are observed in maize, including association with TEs, levels of gene expression and differences in selective constraints.” They propose that the differences observed are attributable to the fact that in maize the different parental genomes were more dissimilar than those in soybean. (Summary by Mary Williams) Plant Cell 10.1105/tpc.17.00595
Many plants are polyploid, meaning that instead of the normal, diploid “2n” complement of chromosomes (one from each parent), they have more than 2n. Following whole-genome duplication, redundancy can allow the duplicated regions to diverge or become silenced. In some cases, silencing occurs preferentially in one of the two parental genomes. By comparing the genomes of maize and soybean to their diploid relatives, Zhao et al. compared the consequences of (ancient) paleopolyploidy events in two species. They found that in maize, “one subgenome has been subject to relaxed selection, lower levels of gene expression, higher rates of transposable element accumulation and more small interfering RNAs and DNA methylation around genes, and higher rates of gene loss,” whereas although soybean does not exhibit subgenome dominance, “members of gene pairs do become differentiated, and that differentiation is associated with several of the same markers of differentiation that are observed in maize, including association with TEs, levels of gene expression and differences in selective constraints.” They propose that the differences observed are attributable to the fact that in maize the different parental genomes were more dissimilar than those in soybean. (Summary by Mary Williams) Plant Cell 10.1105/tpc.17.00595