ZmNAC128 and ZmNAC130 coordinate with Opaque2 to promote endosperm filling in maize
Chen, Yu, He, Peng et al. investigate the synchronized mechanisms of endosperm filling, from nutrient uptake to the biosynthesis of storage reserves.
https://doi.org/10.1093/plcell/koad215
By Di Peng and Zhiyong Zhang
School of Life Sciences, Division of Life Sciences and Medicine, University of Science and Technology of China, Hefei 230027, China
Background: Cereal endosperms store starch and proteins in grains and play a critical role in determining grain yield and quality. Maize (Zea mays), with its sizable grain, is an excellent model for studying cereal endosperm development. Endosperm filling involves two synchronized processes: maternal-to-endosperm nutrient transfer and storage reserve biosynthesis. Several important nutrient transfer-related genes have been identified, and the regulation of storage reserve biosynthesis has been broadly studied in maize. Notably, the transcription factor-encoding genes Opaque2 (O2), ZmNAC128, and ZmNAC130 exhibit strong expression in the filling endosperm. However, how these three transcription factors co-regulate the synchronization of these endosperm-filling processes remains elusive.
Question: What are the roles of ZmNAC128 and ZmNAC130 in endosperm filling? How do they function together with O2 in this process?
Findings: ZmNAC128 and ZmNAC130 directly regulate the expression of all γ-zein genes (encoding storage proteins) and multiple important starch metabolism genes, making them pivotal coordinators of grain quality and yield. Furthermore, ZmNAC128 and ZmNAC130 directly regulate the expression of O2, and together these three transcription factors synergistically activate their own expression through autoregulation and physical interactions. ZmNAC128 and ZmNAC130 also regulate the expression of vital transporter genes responsible for facilitating nutrient transfer from the mother plant to the endosperm. This regulatory mechanism enhances nutrient uptake, with O2 playing a supportive role.
Next steps: We plan to identify additional co-regulatory factors involved in endosperm filling to gain a comprehensive understanding of endosperm filling, from nutrient uptake to the biosynthesis of storage reserves.
Reference:
Erwang Chen, Huiqin Yu, Juan He, Di Peng, Panpan Zhu, Shuxing Pan, Xu Wu, Jincang Wang, Chen Ji, Zhenfei Chao, Zhuopin Xu, Yuejin Wu, Daiyin Chao, Yongrui Wu and Zhiyong Zhang (2023). Transcription factors ZmNAC128 and ZmNAC130 coordinate with OPAQUE2 to promote endosperm filling in maize. https://doi.org/10.1093/plcell/koad215


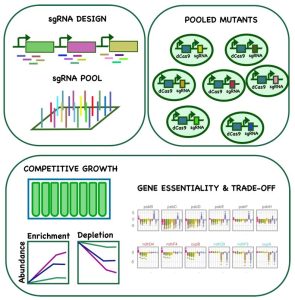 Findings: We screened a large gene repression library consisting of more than 20,000 individual mutants in 11 different conditions. We found that certain genes showed fitness benefits in one condition but were generally detrimental in others, including several genes that play critical roles in mixotrophy and photoheterotrophy, indicating their significance in redirecting metabolic flux. We also discovered genes with condition-specific importance and identified additional functions for central metabolic enzymes. We compiled the fitness data for all genes in all conditions in an interactive, open access web application. In addition, we used our extensive dataset in combination with machine learning to identify “design rules” for effective guide RNAs in the cyanobacterium Synechocystis.
Findings: We screened a large gene repression library consisting of more than 20,000 individual mutants in 11 different conditions. We found that certain genes showed fitness benefits in one condition but were generally detrimental in others, including several genes that play critical roles in mixotrophy and photoheterotrophy, indicating their significance in redirecting metabolic flux. We also discovered genes with condition-specific importance and identified additional functions for central metabolic enzymes. We compiled the fitness data for all genes in all conditions in an interactive, open access web application. In addition, we used our extensive dataset in combination with machine learning to identify “design rules” for effective guide RNAs in the cyanobacterium Synechocystis.