SKIPping over JAZs to improve seed maturation and viability
Vishal Varshney and Manoj Majee, National Institute of Plant Genome Research (NIPGR), Aruna Asaf Ali Marg, New Delhi 110067, India
https://doi.org/10.1093/plcell/koad199
Vishal Varshney et al. investigate the F-box protein SKIP31 in Arabidopsis, revealing a role for this protein in seed maturation, with potential applications in modulating seed vigor.
Background: Seed maturation is a highly complex physiological program during which seeds acquire adaptive features such as seed desiccation tolerance and deposition of seed storage reserves. These features allow seeds to extend their viability and germinability while remaining in a desiccated state for long periods of time. To achieve this feat, seeds engage multi-layered regulatory networks that activate many genes involved in various mechanisms that ultimately improve seed survival and vigor upon maturity. Among these networks, the phytohormone abscisic acid (ABA) and ABA-responsive master regulators play a key role in modulating seed maturation.
Question: The regulatory role of Jasmonate ZIM domain (JAZ) proteins was recently reported in repressing ABA signaling by suppressing ABSCISIC ACID INSENSITIVE3 (ABI3) and ABI5 transcriptional activity during germination. However, how ABI-related transcription factors are repressed by JAZ and how the ABA signaling is de-repressed during seed maturation is unclear.
Findings: We identified the SKIP31–JAZ–ABI5 module as a regulator of seed maturation and seed vigor in Arabidopsis. We show that SKIP31, an F box protein, targets JAZ proteins for proteasomal degradation in a jasmonate (JA)-isoleucine (Ile)-independent manner to alleviate the inhibition imposed by JAZ proteins on ABI5. ABA-mediated downstream signaling thus becomes activated, which is essential for seed maturation, desiccation tolerance and establishment of seed vigor and viability.
Next step: We will test whether other transcription factors and/or regulatory proteins besides ABI5 contribute to the SKIP31–JAZ–ABI5 module to regulate seed maturation. Additionally, we will ask if SKIP31 influences JA-Ile-dependent regulation of the JAZ–MYC module in the JA signaling pathway and JA responses.
Reference:
Vishal Varshney, Abhijit Hazra, Venkateswara Rao, Shraboni Ghosh, Nitin Uttam Kamble, Rakesh Kumar Achary, Shikha Gautam, Manoj Majee (2023) The Arabidopsis F-box protein SKIP31 modulates seed maturation and seed vigor by targeting JAZ proteins independently of jasmonic acid-isoleucine. https://doi.org/10.1093/plcell/koad199


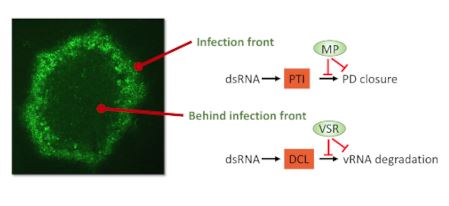 Next steps: This study calls upon the identification of the PTI dsRNA receptor and the mechanisms of PTI signaling (involving identified components such as SERK1, BIK1, calcium channels, CML41, PDLP1/2/3) and PTI suppression by MPs, and how dsRNA-induced PTI and RNA silencing are controlled during the spread of infection.
Next steps: This study calls upon the identification of the PTI dsRNA receptor and the mechanisms of PTI signaling (involving identified components such as SERK1, BIK1, calcium channels, CML41, PDLP1/2/3) and PTI suppression by MPs, and how dsRNA-induced PTI and RNA silencing are controlled during the spread of infection.
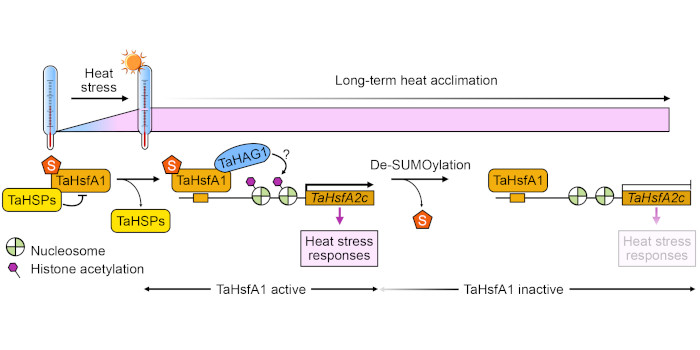 Next steps: We plan to introduce the K459-mutated (non-SUMOylated) form of TaHsfA1 into wheat Tahsfa1 mutants to evaluate the importance of TaHsfA1 SUMOylation in sensing heat stress and regulating responses in cereal crops. In addition, we plan to look for the protein components responsible for TaHsfA1 de-SUMOylation during long-term thermal stress.
Next steps: We plan to introduce the K459-mutated (non-SUMOylated) form of TaHsfA1 into wheat Tahsfa1 mutants to evaluate the importance of TaHsfA1 SUMOylation in sensing heat stress and regulating responses in cereal crops. In addition, we plan to look for the protein components responsible for TaHsfA1 de-SUMOylation during long-term thermal stress.