A cell surface player required for dehydration signaling in foxtail millet
Zhao, Zhang, Liu et al. identify an essential kinase for foxtail millet dehydration tolerance.
https://doi.org/10.1093/plcell/koad200
By Yuxue Zhang and Meicheng Zhao
Key Laboratory of Agricultural Water Resources, Hebei Key Laboratory of Soil Ecology, Center for Agricultural Resources Research, Institute of Genetics and Developmental Biology, Chinese Academy of Sciences, Shijiazhuang, 050021, China
Background: To protect themselves from drought-induced damage, plants must sense the osmotic stress that accompanies drought and rapidly transmit a signal, triggering defense responses to acclimate to water deficit. When plants sense dehydration stress, SNF1-RELATED PROTEIN KINASE2 (SnRK2) family members are activated, representing a key event in dehydration signaling. Although this step was elucidated ~20 years ago, the upstream components that activate SnRK2 kinases remain unknown. We previously identified the transmembrane kinase DROOPY LEAF1 (DPY1) as a key regulator of plant architecture in foxtail millet (Setaria italica). A screen for DPY1-interacing proteins identified a member of the SnRK2 family, suggesting that DPY1 might be involved in SnRK2-mediated dehydration signaling.
Question: Is DPY1 an upstream component required for SnRK2 activation in response to dehydration stress? As a plasma membrane–anchored receptor-like kinase, how does DPY1 respond to dehydration stress?
Findings: DPY1 is crucial for plant acclimation to drought stress. Loss of DPY1 function enhanced susceptibility to drought, partially due to impaired osmotic signaling. DPY1 is phosphorylated and activated in response to osmotic stress and is required for over 50% of osmotic stress–triggered global phosphorylation events, including that of SnRK2s, the central kinases in osmotic stress. DPY1 interacts with but cannot directly phosphorylate STRESS-ACTIVATED PROTEIN KINASE6 (SAPK6), a subclass I SnRK2, but it is required for full SAPK6 activation and the regulation of downstream genes. This activation is largely independent of DPY1-mediated brassinosteroid signaling. Therefore, DPY1 is a key missing component in osmotic stress signaling that mediates SnRK2 activation when plants encounter drought stress.
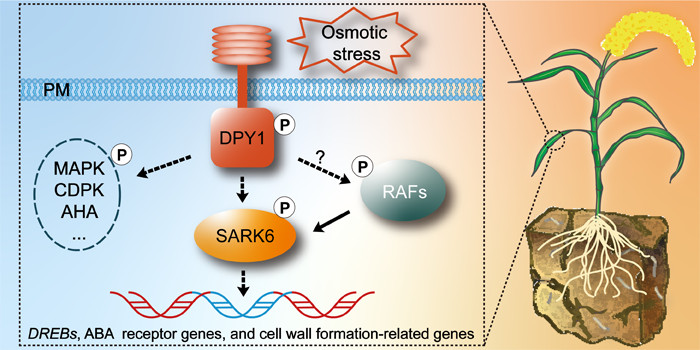
Next steps: Despite the discovery of DPY1-based osmotic stress signaling, numerous gaps remain to be addressed. We plan to focus on identifying the mechanism of DPY1 activation by osmotic stress and components linking DPY1 and SnRK2s.
Reference:
Meicheng Zhao, Qi Zhang, Hong Liu, Sha Tang, Chunyue Shang, Wei Zhang, Yi Sui, Yuxue Zhang, Chunyan Zheng, Hui Zhang, Cuimei Liu, Jinfang Chu, Guanqing Jia, Haigang Wang, Xigang Liu, Diaoguo An, Feng Zhu, Hui Zhi, Chuanyin Wu and Xianmin Diao. (2023). The osmotic stress–activated receptor-like kinase DPY1 mediates SnRK2 kinase activation and drought tolerance in Setariahttps://doi.org/10.1093/plcell/koad200