Overcoming barriers to bioengineering new disease resistance
Bentham, De la Concepcion et al. fine-tuned a plant immune receptor pair to allow for engineering new specificities while avoiding autoactivation.
https://doi.org/10.1093/plcell/koad204
Adam R Bentham, Mark J Banfield
Department of Biochemistry and Metabolism, John Innes Centre, Norwich Research Park, Norwich, NR4 7UH, UK
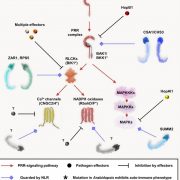
Background: Plants use specialized intracellular immune receptors called nucleotide-binding leucine-rich repeat (NLR) to detect pathogen effector proteins secreted into the host during infection. Effectors aid plant colonization, which can result in serious disease and limit crop yields in agriculture. Therefore, immune responses triggered by NLRs upon perception of effectors are essential to maintain plants healthy. However, plant pathogens use hundreds of effectors during infection and these effectors evolve rapidly, escaping from recognition by NLR immune proteins. Recently, bioengineering of NLRs to recognize a wider range of effectors was demonstrated to be an effective means of generating disease resistant plants. However, bioengineering of NLRs can result in ‘autoactivity’ where the immune system is constantly active. This is deleterious to the health of the plant.
Question: In our study, we strived to understand the limits of bioengineering for a pair of rice NLRs called Pik. Pik are interesting NLRs as they comprise a sensor NLR (important for effector recognition) and a helper NLR (important for defense signaling). Findings: We found bioengineering of the native Pik sensor NLR often resulted in autoactivity. However, by using different combinations of Pik helper NLR alleles (an alternative form of the gene) we could mitigate the autoactivity caused by bioengineering, allowing us to generate Pik sensor NLRs with different effector recognition specificities in the model plant Nicotiana benthamiana. Our study establishes a strategy to incorporate a wider variety of effector recognition modules into the Pik NLRs without autoactivity.
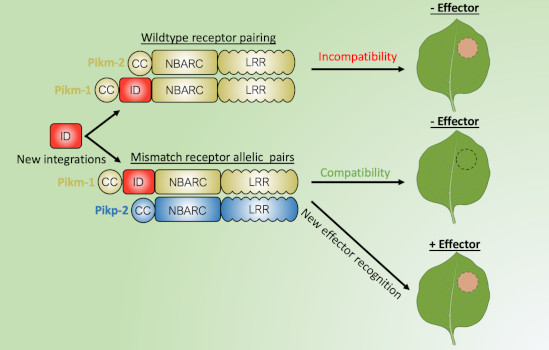
Next Steps: The next step in this research is to understand whether this bioengineered resistance is transferable from model plants to stable transgenic crops, such as rice (Oryza sativa), barley (Hordeum vulgare), wheat (Triticum aestivum) or maize (Zea mays), and to understand which pathogens are the best targets for new disease resistance using the Pik NLRs.
Reference:
Adam R. Bentham, Juan Carlos De la Concepcion, Javier Vega Benjumea, Jiorgos Kourelis, Sally Jones, Melanie Mendel, Jack Stubbs, Clare E. M. Stevenson, Josephine H.R. Maidment, Mark Youles, Rafał Zdrzałek, Sophien Kamoun, Mark J. Banfield (2023) Allelic compatibility in plant immune receptors facilitates engineering of new effector recognition specificities. https://doi.org/10.1093/plcell/koad204