Review: Chloroplast ion homeostasis
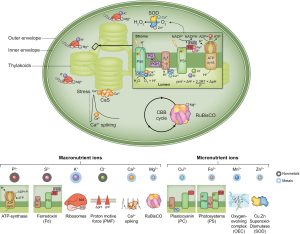 Healthy plants require access to several mineral nutrients, which are usually taken up in ionic form. The details of nutrient uptake, distribution, and function have been painstakingly revealed over several decades. In this excellent new Tansley Review, Kunz et al. provide an overview of ion homeostasis with a focus on the chloroplast and the roles of these mineral ions in photosynthesis. Moving charged ions across membranes requires transport proteins, and plastids have three sets of membranes (outer, inner, and thylakoid) so there is a lot of information about the many transporters involved. Functionally, plastid mineral ions have diverse roles. Several, such as Cu2+, Fe2+, Mn2+, and Zn2+, are necessary as cofactors for the light harvesting and electron transport proteins, and the mechanisms that ensure their homeostasis (right place, right time, right amount) are described. The authors also look at how changing light conditions affect activities of some of the key transporters such as KEA3. They also present the phylogenetic distribution of orthologues across the photosynthetic lineages, noting that cyanobacteria, ancestral plastids, show little overlap with the eukaryotic transportome. The review concludes with a discussion of future research areas, noting that plastid ion homeostasis provides an untapped approach to engineer improved photosynthesis in plants. (Summary Mary Williams @PlantTeaching) New Phytol. 10.1111/nph.19661
Healthy plants require access to several mineral nutrients, which are usually taken up in ionic form. The details of nutrient uptake, distribution, and function have been painstakingly revealed over several decades. In this excellent new Tansley Review, Kunz et al. provide an overview of ion homeostasis with a focus on the chloroplast and the roles of these mineral ions in photosynthesis. Moving charged ions across membranes requires transport proteins, and plastids have three sets of membranes (outer, inner, and thylakoid) so there is a lot of information about the many transporters involved. Functionally, plastid mineral ions have diverse roles. Several, such as Cu2+, Fe2+, Mn2+, and Zn2+, are necessary as cofactors for the light harvesting and electron transport proteins, and the mechanisms that ensure their homeostasis (right place, right time, right amount) are described. The authors also look at how changing light conditions affect activities of some of the key transporters such as KEA3. They also present the phylogenetic distribution of orthologues across the photosynthetic lineages, noting that cyanobacteria, ancestral plastids, show little overlap with the eukaryotic transportome. The review concludes with a discussion of future research areas, noting that plastid ion homeostasis provides an untapped approach to engineer improved photosynthesis in plants. (Summary Mary Williams @PlantTeaching) New Phytol. 10.1111/nph.19661
Unraveling the role of tRNA thiolation in translation control for plant immunity
 Thiolation of tRNA adds a sulfur-containing nucleotide modification at the tRNA wobble position, which is necessary for efficient translation. Zheng et al. investigated the role of tRNA thiolation in plant immunity, revealing its necessity for robust defense responses in Arabidopsis. They identified a pathogen hyper-susceptible mutant, cgb, that encodes ROL5, an enyme required for tRNA thiolation. They showed that, as in yeast, ROL5 interacts with CTU2, which also displays a hyper-susceptible phenotype when function is lost. Using these mutants and proteins, the authors elucidated the molecular mechanisms of tRNA thiolation underlying plant defense. Inhibiting tRNA thiolation causes disruptions in transcriptome and proteome dynamics, particularly impacting NPR1 translation, which highlights the regulatory function of tRNA thiolation in defense signaling pathways. Overall, this research advances our understanding of plant immunity and unveils a novel biological function of tRNA thiolation. (Summary by Yueh Cho @YuehCho1984) eLife. 10.7554/eLife.93517.
Thiolation of tRNA adds a sulfur-containing nucleotide modification at the tRNA wobble position, which is necessary for efficient translation. Zheng et al. investigated the role of tRNA thiolation in plant immunity, revealing its necessity for robust defense responses in Arabidopsis. They identified a pathogen hyper-susceptible mutant, cgb, that encodes ROL5, an enyme required for tRNA thiolation. They showed that, as in yeast, ROL5 interacts with CTU2, which also displays a hyper-susceptible phenotype when function is lost. Using these mutants and proteins, the authors elucidated the molecular mechanisms of tRNA thiolation underlying plant defense. Inhibiting tRNA thiolation causes disruptions in transcriptome and proteome dynamics, particularly impacting NPR1 translation, which highlights the regulatory function of tRNA thiolation in defense signaling pathways. Overall, this research advances our understanding of plant immunity and unveils a novel biological function of tRNA thiolation. (Summary by Yueh Cho @YuehCho1984) eLife. 10.7554/eLife.93517.
How do plants export brassinosteroids?
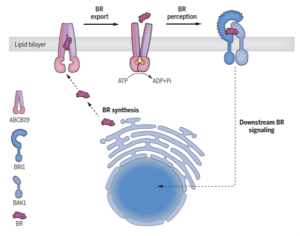 If you’ve ever wondered how plants grow, survive, and adapt to their dynamic environment, the secret lies in their vast array of chemical messengers, also called phytohormones. Brassinosteroids are important hormones that are crucial for plant development and defense against environmental stresses. Like other hormones, brassinosteroids also require specific proteins and other helpers for their perception and signaling inside the cell. Although many of the proteins involved in brassinosteroid signaling have been well-characterized, until now its export mechanism was unknown. In this study, Ying et al. identified ABCB19, an ABC transporter, as an exporter of this hormone. The authors demonstrated the stimulation of ATPase activity of the ABCB19 protein in presence of brassinolides (BL), which confirmed that these proteins interact with BL. Interestingly, this effect was specific to BL. The role of ABCB19 as a brassinosteroid transporter was further established by studying accumulation of BL over time in proteoliposomes containing ABCB19 and Arabidopsis protoplasts incubated with castasterone, a transport substrate for ABCB19. Additionally, in silico studies revealed that the ABCB19 consists of two transmembrane domains, one of which is important for binding with the brassinosteroid molecule. Furthermore, ABCB19 was shown to regulate brassinosteroid signaling together with ABCB1 by using plants with defective ABCB1 and ABCB19 genes. Thus, this study sheds light on an important player involved in transporting brassinosteroids across the cell membrane. (Summary by Abira Sahu @AbiraSahu) Science 10.1126/science.adj4591
If you’ve ever wondered how plants grow, survive, and adapt to their dynamic environment, the secret lies in their vast array of chemical messengers, also called phytohormones. Brassinosteroids are important hormones that are crucial for plant development and defense against environmental stresses. Like other hormones, brassinosteroids also require specific proteins and other helpers for their perception and signaling inside the cell. Although many of the proteins involved in brassinosteroid signaling have been well-characterized, until now its export mechanism was unknown. In this study, Ying et al. identified ABCB19, an ABC transporter, as an exporter of this hormone. The authors demonstrated the stimulation of ATPase activity of the ABCB19 protein in presence of brassinolides (BL), which confirmed that these proteins interact with BL. Interestingly, this effect was specific to BL. The role of ABCB19 as a brassinosteroid transporter was further established by studying accumulation of BL over time in proteoliposomes containing ABCB19 and Arabidopsis protoplasts incubated with castasterone, a transport substrate for ABCB19. Additionally, in silico studies revealed that the ABCB19 consists of two transmembrane domains, one of which is important for binding with the brassinosteroid molecule. Furthermore, ABCB19 was shown to regulate brassinosteroid signaling together with ABCB1 by using plants with defective ABCB1 and ABCB19 genes. Thus, this study sheds light on an important player involved in transporting brassinosteroids across the cell membrane. (Summary by Abira Sahu @AbiraSahu) Science 10.1126/science.adj4591
Methyltransferase TaSAMT1 mediates wheat freezing tolerance by integrating brassinosteroid and salicylic acid signaling
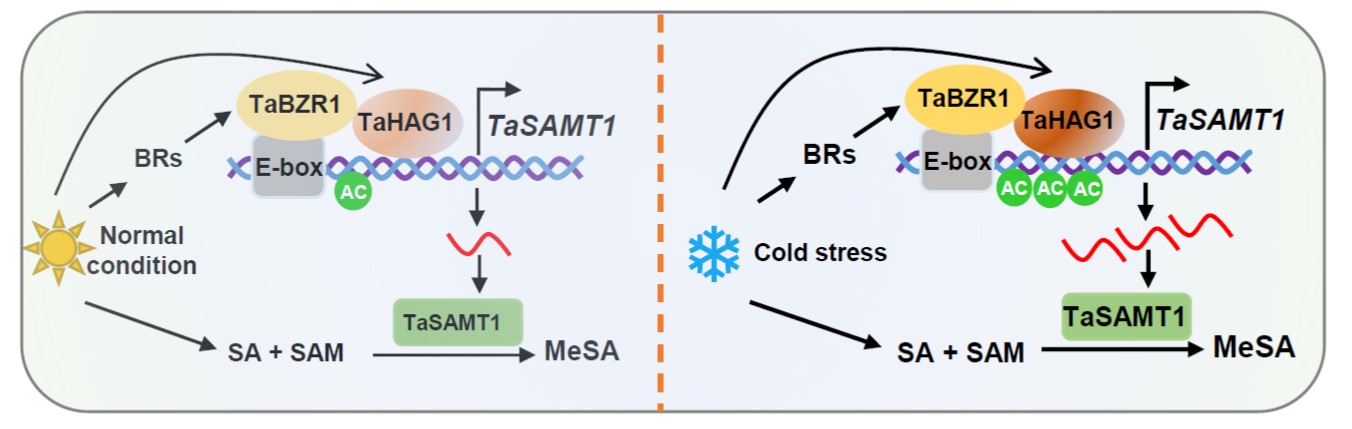 Temperature extremes, such as cold stress, severely affect wheat (Triticum aestivum) productivity and quality by impairing its vegetative and reproductive growth. Several phytohormones have roles in cold stress, such as brassinosteroids (BRs) and salicylic acid (SA). However, how BR interacts with SA in response to cold stress remains unknown. In this work, Chu et al. describe the role of an SA methyltransferase named TaSAMT1 and its function in conferring freezing tolerance in wheat. They demonstrated that TaSAMT1 converts SA to methyl SA (MeSA), a volatile form of SA that acts as a signal molecule in plant responses to abiotic stress. Through various experiments, including use of Arabidopsis thaliana knock-out and overexpressing mutant lines, they established a role of MeSA in the response to freezing tolerance in wheat. In addition, they elucidated how SA and BRs pathways interact in the regulation of freezing tolerance. Specifically, they found that BRASSINAZOLE-RESISTANT 1 (TaBZR1) directly interacts with the TaSAMT1 promoter, inducing its transcription. Finally, TaBZR1 also interacts with TaHAG1, a histone acetyltransferase that enhances TaSAMT1 expression via histone acetylation but it also interacts with TaHAG1, a histone acetyltransferase that enhances TaSAMT1 expression via histone acetylation. This work suggests that TaSAMT1 might be a target for selection and breeding for improving wheat cold tolerance. (Summary by Eva Maria Gomez Alvarez @eva_ga96) Plant Cell 10.1093/plcell/koae100
Temperature extremes, such as cold stress, severely affect wheat (Triticum aestivum) productivity and quality by impairing its vegetative and reproductive growth. Several phytohormones have roles in cold stress, such as brassinosteroids (BRs) and salicylic acid (SA). However, how BR interacts with SA in response to cold stress remains unknown. In this work, Chu et al. describe the role of an SA methyltransferase named TaSAMT1 and its function in conferring freezing tolerance in wheat. They demonstrated that TaSAMT1 converts SA to methyl SA (MeSA), a volatile form of SA that acts as a signal molecule in plant responses to abiotic stress. Through various experiments, including use of Arabidopsis thaliana knock-out and overexpressing mutant lines, they established a role of MeSA in the response to freezing tolerance in wheat. In addition, they elucidated how SA and BRs pathways interact in the regulation of freezing tolerance. Specifically, they found that BRASSINAZOLE-RESISTANT 1 (TaBZR1) directly interacts with the TaSAMT1 promoter, inducing its transcription. Finally, TaBZR1 also interacts with TaHAG1, a histone acetyltransferase that enhances TaSAMT1 expression via histone acetylation but it also interacts with TaHAG1, a histone acetyltransferase that enhances TaSAMT1 expression via histone acetylation. This work suggests that TaSAMT1 might be a target for selection and breeding for improving wheat cold tolerance. (Summary by Eva Maria Gomez Alvarez @eva_ga96) Plant Cell 10.1093/plcell/koae100
A nitrogen-fixing algal organelle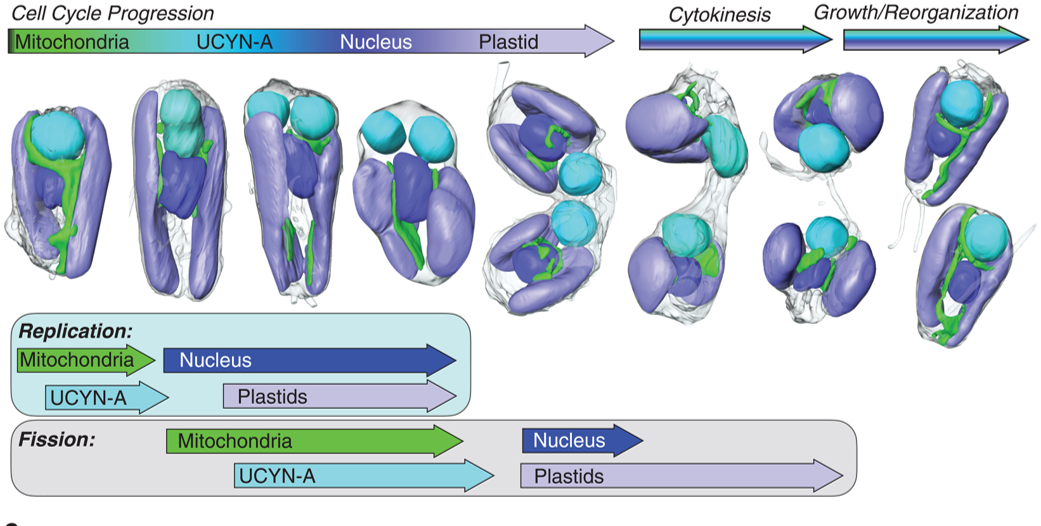 Coale et al. provide an exciting peek into the evolution of a nitrogen-fixing organelle (called a nitroplast) in their studies of a tiny marine alga, Braarudosphaera bigelowii and its endosymbiont cyanobacterium, Candidatus Atelocyanobacterium thalassa (UCYN-A). Plant biologists are familiar with other nitrogen-fixing endosymbionts, such as the rhizobia that reside in plant nodules. And of course, we are all familiar with the mitochondria and plastids, descendants of ancient endosymbionts that have lost their ability to live independently of the host cell. The interesting new findings presented by Coale et al. suggest that the UCYN-A, long-considered to be an endosymbiont, has evolved into an organelle. The authors used soft x-ray tomography to image the single-celled alga during division. They found that the single endosymbiont/nitroplast in each cell replicates alongside the other cellular compartments, and one each of the resulting structures is passed into each daughter cell. What cements this as an organelle is that many genes required for its viability and function (such as genes required to synthesize amino acids) are encoded in the host genome and imported into the nitroplast. Interestingly, these proteins are marked by a novel transit peptide that directs them to the nitroplast, analogous to how nuclear-encoded plastid or mitochondrial proteins are imported. Besides providing yet another example of how “life finds a way”, this study opens the door to the possibility of introducing nitroplasts into crop plants and alleviating a major agricultural challenge. (Summary by Mary Williams @PlantTeaching) Science 10.1126/science.adk1075
Coale et al. provide an exciting peek into the evolution of a nitrogen-fixing organelle (called a nitroplast) in their studies of a tiny marine alga, Braarudosphaera bigelowii and its endosymbiont cyanobacterium, Candidatus Atelocyanobacterium thalassa (UCYN-A). Plant biologists are familiar with other nitrogen-fixing endosymbionts, such as the rhizobia that reside in plant nodules. And of course, we are all familiar with the mitochondria and plastids, descendants of ancient endosymbionts that have lost their ability to live independently of the host cell. The interesting new findings presented by Coale et al. suggest that the UCYN-A, long-considered to be an endosymbiont, has evolved into an organelle. The authors used soft x-ray tomography to image the single-celled alga during division. They found that the single endosymbiont/nitroplast in each cell replicates alongside the other cellular compartments, and one each of the resulting structures is passed into each daughter cell. What cements this as an organelle is that many genes required for its viability and function (such as genes required to synthesize amino acids) are encoded in the host genome and imported into the nitroplast. Interestingly, these proteins are marked by a novel transit peptide that directs them to the nitroplast, analogous to how nuclear-encoded plastid or mitochondrial proteins are imported. Besides providing yet another example of how “life finds a way”, this study opens the door to the possibility of introducing nitroplasts into crop plants and alleviating a major agricultural challenge. (Summary by Mary Williams @PlantTeaching) Science 10.1126/science.adk1075
Altering root system architecture in barley without impacting above-ground traits
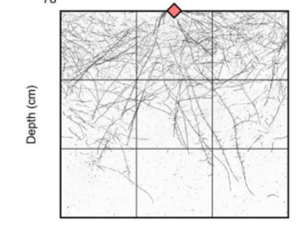 Roots are important for acquiring water and nutrients from the soil; however root system architecture is poorly understood in cereal crops. Here Aldiss et al. used CRISPR/Cas9 to generate barley mutants in the auxin transporter PIN-FORMED2 (PIN2). Seedlings were grown in chambers and after four days the chambers were rotated by 90o. Whilst the roots of wild type plants grew towards the gravity vector, the roots of mutant plants did not, suggesting they were agravitropic. To look at the root system architecture of more mature plants, the mutants were grown in wide rhizoboxes for 28 days. Analysis of the root system using the image analysis software RhizoVision Explorer showed that pin2 plants had a wider root system and more shallow roots, defined as having a root angle of less than 30o. Interestingly, pin2 plants had no significant difference in shoot biomass and no differences in either tiller number or time to flowering. Hence mutating PIN2 provides a way of generating a wide, shallow root system in barley without compromising above ground traits, which could be beneficial for capturing phosphorus or water from the topsoil. (Summary by Rose McNeilly @Rose_McN) bioRxiv 10.1101/2024.03.28.587117v1
Roots are important for acquiring water and nutrients from the soil; however root system architecture is poorly understood in cereal crops. Here Aldiss et al. used CRISPR/Cas9 to generate barley mutants in the auxin transporter PIN-FORMED2 (PIN2). Seedlings were grown in chambers and after four days the chambers were rotated by 90o. Whilst the roots of wild type plants grew towards the gravity vector, the roots of mutant plants did not, suggesting they were agravitropic. To look at the root system architecture of more mature plants, the mutants were grown in wide rhizoboxes for 28 days. Analysis of the root system using the image analysis software RhizoVision Explorer showed that pin2 plants had a wider root system and more shallow roots, defined as having a root angle of less than 30o. Interestingly, pin2 plants had no significant difference in shoot biomass and no differences in either tiller number or time to flowering. Hence mutating PIN2 provides a way of generating a wide, shallow root system in barley without compromising above ground traits, which could be beneficial for capturing phosphorus or water from the topsoil. (Summary by Rose McNeilly @Rose_McN) bioRxiv 10.1101/2024.03.28.587117v1
All fruit things come to an end: FRUITFULL controls end of flowering and seed yield in pea
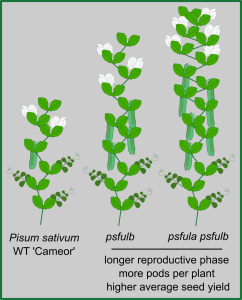 The onset of flowering is a tightly regulated process, as is the end of the reproductive phase in plants. In Arabidopsis thaliana, FRUITFULL (FUL) contributes to ending the reproductive cycle, as do signals derived from developing seeds. Here, Martínez-Fernández et al. explored to which extent that role extends to other plant species, namely pea (Pisum sativum ‘Cameor’). The expression levels of pea FUL genes PsFULa and PsFULb are upregulated upon floral transition. As in Arabidopsis, loss-of-function mutant psfulb and double-mutant psfula/b lines have longer reproductive cycles and develop more pods and seeds per plant than WT. Fruit and seed morphology and composition, however, do not differ significantly between WT and mutant lines. As seed-derived signals also contribute to ending the reproductive phase, the authors investigated the interaction between the FUL genes and seed signals. When seed formation was inhibited, all plants had longer reproductive phases, with the extension in psfulb and psfula/b phases approximately 9- and 12-fold longer, respectively. This shows that the FUL genes and seed-derived signals synergistically determine the end of the reproductive phase. Field trials revealed that psfulb and psfula/b plants yield significantly higher seed production when compared to WT, showcasing the agricultural potential that might be harnessed by understanding end-of-flowering regulation. (Summary by John Vilasboa @vilasjohn) Proc Natl Acad Sci. USA 10.1073/pnas.2321975121
The onset of flowering is a tightly regulated process, as is the end of the reproductive phase in plants. In Arabidopsis thaliana, FRUITFULL (FUL) contributes to ending the reproductive cycle, as do signals derived from developing seeds. Here, Martínez-Fernández et al. explored to which extent that role extends to other plant species, namely pea (Pisum sativum ‘Cameor’). The expression levels of pea FUL genes PsFULa and PsFULb are upregulated upon floral transition. As in Arabidopsis, loss-of-function mutant psfulb and double-mutant psfula/b lines have longer reproductive cycles and develop more pods and seeds per plant than WT. Fruit and seed morphology and composition, however, do not differ significantly between WT and mutant lines. As seed-derived signals also contribute to ending the reproductive phase, the authors investigated the interaction between the FUL genes and seed signals. When seed formation was inhibited, all plants had longer reproductive phases, with the extension in psfulb and psfula/b phases approximately 9- and 12-fold longer, respectively. This shows that the FUL genes and seed-derived signals synergistically determine the end of the reproductive phase. Field trials revealed that psfulb and psfula/b plants yield significantly higher seed production when compared to WT, showcasing the agricultural potential that might be harnessed by understanding end-of-flowering regulation. (Summary by John Vilasboa @vilasjohn) Proc Natl Acad Sci. USA 10.1073/pnas.2321975121
SWEET sugar transporters orchestrate distribution of microbiota along the longitudinal axis of Arabidopsis roots
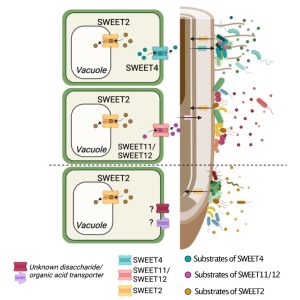 Plant roots are functionally distinct along the longitudinal axis due to different cell types and diverse metabolic states. Root-secreted metabolites are involved in the assembly of complex microbial communities, yet the relationships between root-metabolites and organization of root microbiota at the spatial scale is poorly understood. This is partly because of a lack of suitable system to study such relationships by simultaneous profiling of metabolites and microbiota along the longitudinal axis of roots. Loo et al. developed two complementary plant growth systems, CD-rhizotron and ArtSoil, to study such relationships. Through bacterial community profiling (root endospheric and rhizosphere fractions) and metabolomics analyses, they showed that differential microbial community structure coincided with a differential metabolite accumulation along the longitudinal axis, suggesting spatial metabolite profiles could be implicated for orchestrating the microbial community. Further, applying bioinformatics, GUS histochemical analyses, and Arabidopsis mutant lines, they discovered a relationship between SWEET transporters (sugar transporters) and microbial communities along the longitudinal root axes. Specifically, several SWEET uniporters were induced by microbiota along the longitudinal root axis, and they contributed to microbial community assembly. This study established the spatial coordination between sugar transporters and microbial community assembly in roots. (Summary by Arijit Mukherjee @ArijitM61745830) Cell Host Microbe 10.1016/j.chom.2024.02.014
Plant roots are functionally distinct along the longitudinal axis due to different cell types and diverse metabolic states. Root-secreted metabolites are involved in the assembly of complex microbial communities, yet the relationships between root-metabolites and organization of root microbiota at the spatial scale is poorly understood. This is partly because of a lack of suitable system to study such relationships by simultaneous profiling of metabolites and microbiota along the longitudinal axis of roots. Loo et al. developed two complementary plant growth systems, CD-rhizotron and ArtSoil, to study such relationships. Through bacterial community profiling (root endospheric and rhizosphere fractions) and metabolomics analyses, they showed that differential microbial community structure coincided with a differential metabolite accumulation along the longitudinal axis, suggesting spatial metabolite profiles could be implicated for orchestrating the microbial community. Further, applying bioinformatics, GUS histochemical analyses, and Arabidopsis mutant lines, they discovered a relationship between SWEET transporters (sugar transporters) and microbial communities along the longitudinal root axes. Specifically, several SWEET uniporters were induced by microbiota along the longitudinal root axis, and they contributed to microbial community assembly. This study established the spatial coordination between sugar transporters and microbial community assembly in roots. (Summary by Arijit Mukherjee @ArijitM61745830) Cell Host Microbe 10.1016/j.chom.2024.02.014
Unlocking nature’s arsenal: Engineering grasses for insect defense and livestock palatability
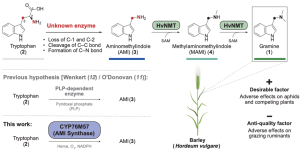 The defensive alkaloid gramine, present in barley and other grass species, plays an important role in protecting the plant from insect damage but poses challenges for ruminant palatability. Breeding strategies balance these factors by maintaining the protective function and making the grain palatable for grazing animals. In this study, Dias et al. used pan-genome sequencing to identify a gramine biosynthesis gene cluster in barley. They further characterized an enzyme responsible for the intricate oxidative rearrangement of tryptophan. Through the adjustment of gramine levels in yeast and various plant species, the authors demonstrated that gramine-related traits could be optimized. This study represents a big step forward in understanding grass defenses for agricultural improvement. (Summary by Yueh Cho @YuehCho1984) Science. 10.1126/science.adk6112.
The defensive alkaloid gramine, present in barley and other grass species, plays an important role in protecting the plant from insect damage but poses challenges for ruminant palatability. Breeding strategies balance these factors by maintaining the protective function and making the grain palatable for grazing animals. In this study, Dias et al. used pan-genome sequencing to identify a gramine biosynthesis gene cluster in barley. They further characterized an enzyme responsible for the intricate oxidative rearrangement of tryptophan. Through the adjustment of gramine levels in yeast and various plant species, the authors demonstrated that gramine-related traits could be optimized. This study represents a big step forward in understanding grass defenses for agricultural improvement. (Summary by Yueh Cho @YuehCho1984) Science. 10.1126/science.adk6112.
Hidden influence: How microbial stress responses shape plant natural selection
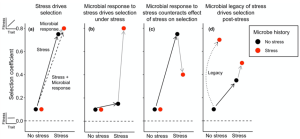 Soil microbial communities respond rapidly to stress, potentially leading to altered compositions in stressful environments and consequently impacting plant natural selection. Bolin and Lau investigated the influence of microbial responses to stress and persistent microbial legacy effects on plant selection. Chamaecrista fasciculata (partridge pea) plants were cultivated under stressful (salt, herbicide, or herbivory) or non-stressful conditions with microbes previously exposed to these environments. Microbial responses to stress counteracted the direct effects of stress on plant selection, thereby attenuating stress’s role as a selective agent. Additionally, microbial legacy effects of stress altered plant selection in non-stressful environments, indicating a prolonged impact beyond the stress period. These findings suggest that soil microbes may alter plant adaptation to stress, potentially reshaping the evolutionary trajectory of plant populations by reducing stress’s selective pressure. (Summary by Yueh Cho @YuehCho1984) New Phytologist. 10.1111/nph.19707.
Soil microbial communities respond rapidly to stress, potentially leading to altered compositions in stressful environments and consequently impacting plant natural selection. Bolin and Lau investigated the influence of microbial responses to stress and persistent microbial legacy effects on plant selection. Chamaecrista fasciculata (partridge pea) plants were cultivated under stressful (salt, herbicide, or herbivory) or non-stressful conditions with microbes previously exposed to these environments. Microbial responses to stress counteracted the direct effects of stress on plant selection, thereby attenuating stress’s role as a selective agent. Additionally, microbial legacy effects of stress altered plant selection in non-stressful environments, indicating a prolonged impact beyond the stress period. These findings suggest that soil microbes may alter plant adaptation to stress, potentially reshaping the evolutionary trajectory of plant populations by reducing stress’s selective pressure. (Summary by Yueh Cho @YuehCho1984) New Phytologist. 10.1111/nph.19707.
Unintended consequences of planting native and non- native trees in treeless ecosystems to mitigate climate change
 Afforestation initiatives are globally underway to replace naturally treeless ecosystems with native and non-native trees. The primary objective is to enhance atmospheric carbon capture as a means to combat climate change. This review seeks to illuminate the diverse impacts of afforestation, both positive and negative, while also proposing mitigation strategies. Notably, the replacement of grasslands and shrublands during afforestation can adversely affect biodiversity. Negative consequences may include water yield reduction, soil salinization, soil carbon depletion, heightened wildfire risk, and increased soil temperatures. Additionally, the proliferation of woody plants presents a health hazard by facilitating the spread of vector-borne diseases. Given the complexity of afforestation efforts, it is crucial to consider various factors such as tree species selection, location suitability, and the duration of tree growth. Special care is warranted when introducing non-native trees to prevent them from replacing native species. Furthermore, prioritizing conservation and restoration initiatives over afforestation may offer greater potential for carbon sequestration. (Summary by Villő Bernád @BernadVillo) J. Ecol. 10.1111/1365-2745.14300
Afforestation initiatives are globally underway to replace naturally treeless ecosystems with native and non-native trees. The primary objective is to enhance atmospheric carbon capture as a means to combat climate change. This review seeks to illuminate the diverse impacts of afforestation, both positive and negative, while also proposing mitigation strategies. Notably, the replacement of grasslands and shrublands during afforestation can adversely affect biodiversity. Negative consequences may include water yield reduction, soil salinization, soil carbon depletion, heightened wildfire risk, and increased soil temperatures. Additionally, the proliferation of woody plants presents a health hazard by facilitating the spread of vector-borne diseases. Given the complexity of afforestation efforts, it is crucial to consider various factors such as tree species selection, location suitability, and the duration of tree growth. Special care is warranted when introducing non-native trees to prevent them from replacing native species. Furthermore, prioritizing conservation and restoration initiatives over afforestation may offer greater potential for carbon sequestration. (Summary by Villő Bernád @BernadVillo) J. Ecol. 10.1111/1365-2745.14300
 Soil microbial communities respond rapidly to stress, potentially leading to altered compositions in stressful environments and consequently impacting plant natural selection. Bolin and Lau investigated the influence of microbial responses to stress and persistent microbial legacy effects on plant selection. Chamaecrista fasciculata (partridge pea) plants were cultivated under stressful (salt, herbicide, or herbivory) or non-stressful conditions with microbes previously exposed to these environments. Microbial responses to stress counteracted the direct effects of stress on plant selection, thereby attenuating stress’s role as a selective agent. Additionally, microbial legacy effects of stress altered plant selection in non-stressful environments, indicating a prolonged impact beyond the stress period. These findings suggest that soil microbes may alter plant adaptation to stress, potentially reshaping the evolutionary trajectory of plant populations by reducing stress’s selective pressure. (Summary by Yueh Cho @YuehCho1984) New Phytologist. 10.1111/nph.19707.
Soil microbial communities respond rapidly to stress, potentially leading to altered compositions in stressful environments and consequently impacting plant natural selection. Bolin and Lau investigated the influence of microbial responses to stress and persistent microbial legacy effects on plant selection. Chamaecrista fasciculata (partridge pea) plants were cultivated under stressful (salt, herbicide, or herbivory) or non-stressful conditions with microbes previously exposed to these environments. Microbial responses to stress counteracted the direct effects of stress on plant selection, thereby attenuating stress’s role as a selective agent. Additionally, microbial legacy effects of stress altered plant selection in non-stressful environments, indicating a prolonged impact beyond the stress period. These findings suggest that soil microbes may alter plant adaptation to stress, potentially reshaping the evolutionary trajectory of plant populations by reducing stress’s selective pressure. (Summary by Yueh Cho @YuehCho1984) New Phytologist. 10.1111/nph.19707.












 Question: What is the role of microtubule nucleation and geometry in phragmoplast asymmetry?
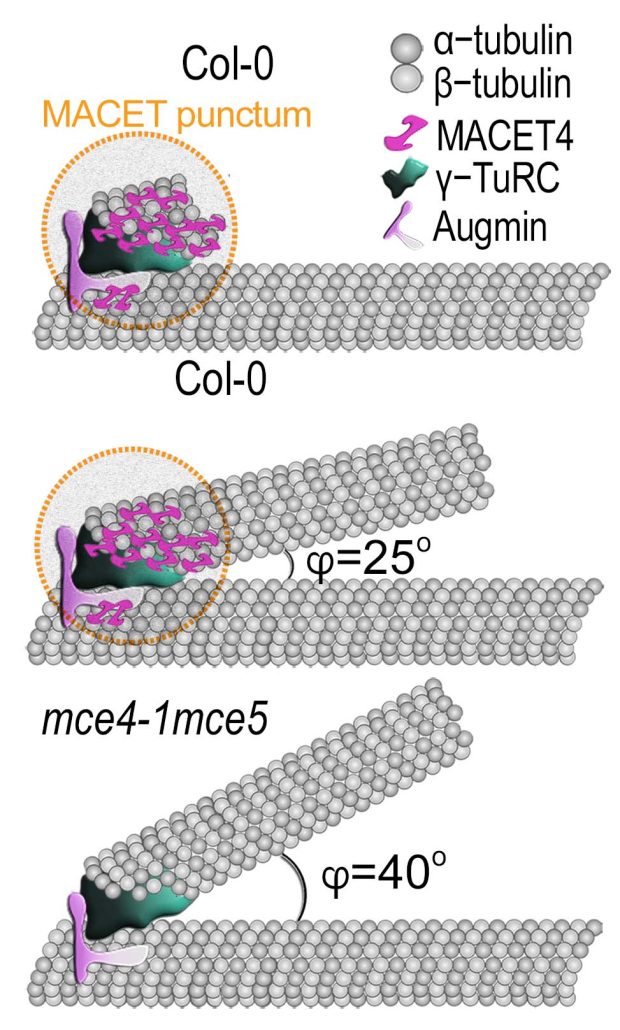
Question: What is the role of microtubule nucleation and geometry in phragmoplast asymmetry? Findings: Knockout of the single type-II metacaspase gene of Chlamydomonas (named CrMCA-II) decreased algal fitness under heat stress (42°C), resulting in lower growth and increased cell death. Unlike other studied plant metacaspases, CrMCA-II was abundantly associated with the plasma membrane but partially translocated to the cytoplasm during heat treatment. Another peculiar feature of CrMCA-II was its oligomerization to form both catalytically active dimers and larger, megadalton-scale complexes comprising inactive pro-enzyme (zymogen). We discovered that the thermoprotective function of CrMCA-II is independent of its proteolytic activity but instead might be associated with an unknown mechanism by which CrMCA-II mediates fluidization of the plasma membrane.
Findings: Knockout of the single type-II metacaspase gene of Chlamydomonas (named CrMCA-II) decreased algal fitness under heat stress (42°C), resulting in lower growth and increased cell death. Unlike other studied plant metacaspases, CrMCA-II was abundantly associated with the plasma membrane but partially translocated to the cytoplasm during heat treatment. Another peculiar feature of CrMCA-II was its oligomerization to form both catalytically active dimers and larger, megadalton-scale complexes comprising inactive pro-enzyme (zymogen). We discovered that the thermoprotective function of CrMCA-II is independent of its proteolytic activity but instead might be associated with an unknown mechanism by which CrMCA-II mediates fluidization of the plasma membrane.