CAM evolution is associated with gene family expansion in an explosive bromeliad radiation
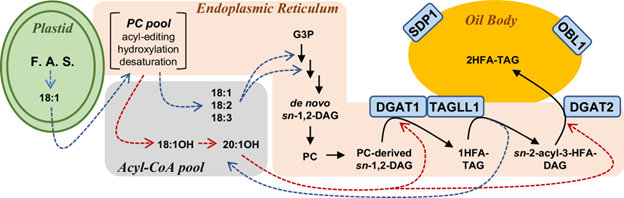
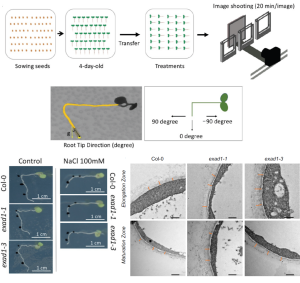
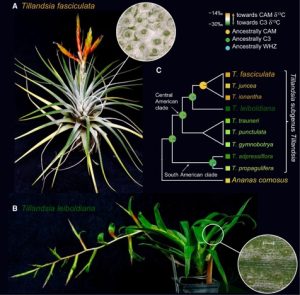 Studies on Crassulacean Acid Metabolism (CAM) plant genomes are scarse and CAM evolution is known to be an ecological driver of diversification. The subgenus Tillandsia (Bromeliaceae) is one of the fastest diversifying clades in the plant kingdom and is known for its adaptive CAM trait. It has recently been subect to comparative genomic studies by Crego et al. CAM evolution is known to be mostly driven by gene family expansion, hence the authors studied two ecologically divergent members of subgenus Tillandsia, Tillandsia fasculata (a CAM plant) and Tillandsia leiboldiana, a C3 plant. De-novo haploid genome assembly of these two species were constructed. Gene family expansion in T. fasciculata was greater than T. leiboldiana in terms of increased number of genes belonging to multicopy families followed by a higher rate of gene duplication. In-depth gene expression analyis on the two genomes also showed differences related to CAM evolution, primarily at the regulatory level. CAM-related genes showed differential expression (DE) between both species across a 24h period. CAM-related DE-gene expression profiles are more aligned to the T. fasciculata genome, possibly leading to its CAM phenotype. Integration of such de novo assembly studies with in-depth transcriptome analysis to understand complex traits like CAM evolution is important in context of evolutionary biology studies on this highly species-rich plant family. (Summary by Indrani Kakati Baruah @Indranik333) Plant Cell 10.1093/plcell/koae130
Studies on Crassulacean Acid Metabolism (CAM) plant genomes are scarse and CAM evolution is known to be an ecological driver of diversification. The subgenus Tillandsia (Bromeliaceae) is one of the fastest diversifying clades in the plant kingdom and is known for its adaptive CAM trait. It has recently been subect to comparative genomic studies by Crego et al. CAM evolution is known to be mostly driven by gene family expansion, hence the authors studied two ecologically divergent members of subgenus Tillandsia, Tillandsia fasculata (a CAM plant) and Tillandsia leiboldiana, a C3 plant. De-novo haploid genome assembly of these two species were constructed. Gene family expansion in T. fasciculata was greater than T. leiboldiana in terms of increased number of genes belonging to multicopy families followed by a higher rate of gene duplication. In-depth gene expression analyis on the two genomes also showed differences related to CAM evolution, primarily at the regulatory level. CAM-related genes showed differential expression (DE) between both species across a 24h period. CAM-related DE-gene expression profiles are more aligned to the T. fasciculata genome, possibly leading to its CAM phenotype. Integration of such de novo assembly studies with in-depth transcriptome analysis to understand complex traits like CAM evolution is important in context of evolutionary biology studies on this highly species-rich plant family. (Summary by Indrani Kakati Baruah @Indranik333) Plant Cell 10.1093/plcell/koae130