Plant Science Research Weekly: May 17, 2024
Review. Revisiting plant electric signaling: Challenging an old phenomenon with new discoveries
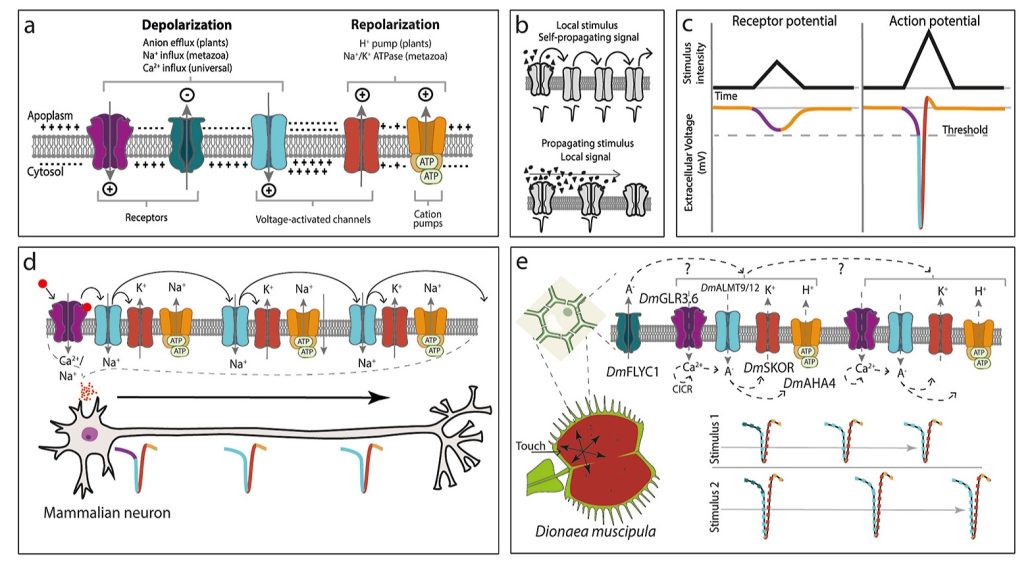 In the electrifying world of plant signaling, a paradigm shift is underway as researchers dig into the intricate mechanisms of action potentials (APs) and slow wave potentials (SWPs). Departing from conventional neurophysiological dogma in the animal kingdom, this review by Barbosa-Caro and Wudick illuminates the nuanced dance of electric signals within plants. Unlike their animal counterparts, plant APs defy easy classification, challenging our understanding of electrical signal transmission. SWPs, with their depolarizing deviations, further complicate the picture, necessitating a holistic approach to unravel their mysteries. From membrane depolarization to calcium fluxes, a symphony of molecular players orchestrates these signals, begging for a tailored conceptual framework. While similarities to neuronal transmission tantalize, the fluctuating dynamics of plant APs hint at a fundamentally different playbook. Yet, amidst the complexity, a beacon of hope emerges: the integration of quantitative modelling and cutting-edge tools promises to illuminate the dark corners of plant electrophysiology. As we embark on this electrifying journey, the field brims with new questions, beckoning us to explore the frontiers of botanical excitement with open minds and nimble tools. (Summary by Nibedita Priyadarshini) Curr. Opin. Plant Biol. 10.1016/j.pbi.2024.102528.
In the electrifying world of plant signaling, a paradigm shift is underway as researchers dig into the intricate mechanisms of action potentials (APs) and slow wave potentials (SWPs). Departing from conventional neurophysiological dogma in the animal kingdom, this review by Barbosa-Caro and Wudick illuminates the nuanced dance of electric signals within plants. Unlike their animal counterparts, plant APs defy easy classification, challenging our understanding of electrical signal transmission. SWPs, with their depolarizing deviations, further complicate the picture, necessitating a holistic approach to unravel their mysteries. From membrane depolarization to calcium fluxes, a symphony of molecular players orchestrates these signals, begging for a tailored conceptual framework. While similarities to neuronal transmission tantalize, the fluctuating dynamics of plant APs hint at a fundamentally different playbook. Yet, amidst the complexity, a beacon of hope emerges: the integration of quantitative modelling and cutting-edge tools promises to illuminate the dark corners of plant electrophysiology. As we embark on this electrifying journey, the field brims with new questions, beckoning us to explore the frontiers of botanical excitement with open minds and nimble tools. (Summary by Nibedita Priyadarshini) Curr. Opin. Plant Biol. 10.1016/j.pbi.2024.102528.
Review: The era of panomics-driven gene discovery in plants
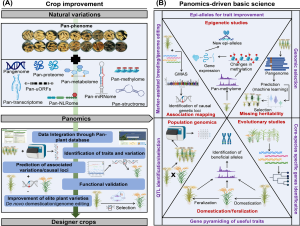 Panomics, an approach integrating multiple ‘omics’ datasets such as genomics, transcriptomics, metabolomics, and phenomics, has seen rapid advancement in recent years due to technological improvements, particularly in genomics. This review focuses on the recent developments in panomics-driven gene discovery and its application in plant breeding and development, particularly for enhancing stress tolerance, specialized metabolite accumulation, and rediscoveries. Pangenomics, which involves representing a species’ genome derived from diverse cultivars, plays a significant role in gene discovery and mining, while recent advances have facilitated the detection of population-wide structural variations. These tools offer great potential for wider application in plant breeding, aiding in the understanding of domestication and facilitating the reintroduction of eliminated advantageous traits, a process known as redomestication. Furthermore, ongoing developments, such as the identification of pan-miRNA and other noncoding RNA, promise to broaden our understanding of miRNA/mRNA regulatory modules. Additionally, panomics addresses the complexities presented by polyploid genomes, accelerates the integration of understudied crops into the genomic era, and bridges the gap between cultivated crops and their wild counterparts. (Summary by Villő Bernád) Trends Plant Sci. 10.1016/j.tplants.2024.03.007
Panomics, an approach integrating multiple ‘omics’ datasets such as genomics, transcriptomics, metabolomics, and phenomics, has seen rapid advancement in recent years due to technological improvements, particularly in genomics. This review focuses on the recent developments in panomics-driven gene discovery and its application in plant breeding and development, particularly for enhancing stress tolerance, specialized metabolite accumulation, and rediscoveries. Pangenomics, which involves representing a species’ genome derived from diverse cultivars, plays a significant role in gene discovery and mining, while recent advances have facilitated the detection of population-wide structural variations. These tools offer great potential for wider application in plant breeding, aiding in the understanding of domestication and facilitating the reintroduction of eliminated advantageous traits, a process known as redomestication. Furthermore, ongoing developments, such as the identification of pan-miRNA and other noncoding RNA, promise to broaden our understanding of miRNA/mRNA regulatory modules. Additionally, panomics addresses the complexities presented by polyploid genomes, accelerates the integration of understudied crops into the genomic era, and bridges the gap between cultivated crops and their wild counterparts. (Summary by Villő Bernád) Trends Plant Sci. 10.1016/j.tplants.2024.03.007
De novo domestication: What about the weedy relatives?
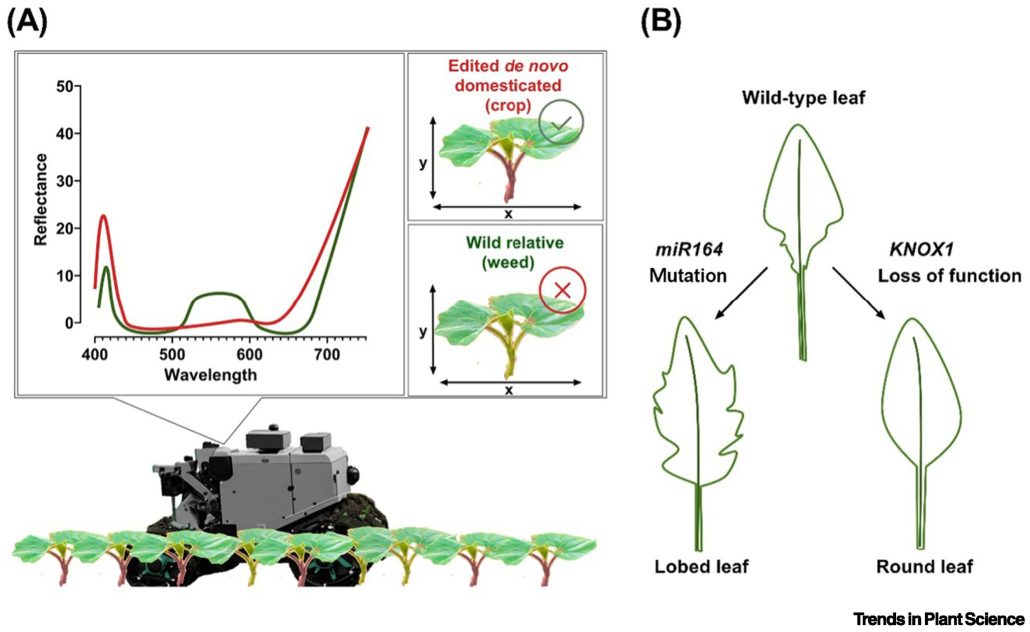 Because much of the genetic diversity of our crops plants was lost during domestication, many breeding efforts endeavor to cross in resilience genes from wild relatives. An alternative approach is to rapidly domesticate those wild relatives themselves, for example by editing genes that affect flowering time, seed shattering, or fruit or seed size. In this interesting Opinion article, Correia et al. raise the point that the wild, undomesticated close relatives of these de novo domesticated varieties are likely to be weedy pests and difficult to eliminate from fields. They propose an innovative solution, which is to also introduce traits into the new varieties that make them easy to spot for a field-based weeding robot. For example, the desired variety could express pigments such as anthocyanins or carotenoids in places the wild relative does not, signaling the robot to leave it alone. Alternatively, leaf shape could be changed from serrated to round, or vice versa. The authors point out that these small but meaningful changes in plant phenotype can be made through genome editing, avoiding the pitfalls of introducing foreign genes. (Summary by Mary Williams @PlantTeaching) Trends Plant Sci. 10.1016/j.tplants.2024.03.001
Because much of the genetic diversity of our crops plants was lost during domestication, many breeding efforts endeavor to cross in resilience genes from wild relatives. An alternative approach is to rapidly domesticate those wild relatives themselves, for example by editing genes that affect flowering time, seed shattering, or fruit or seed size. In this interesting Opinion article, Correia et al. raise the point that the wild, undomesticated close relatives of these de novo domesticated varieties are likely to be weedy pests and difficult to eliminate from fields. They propose an innovative solution, which is to also introduce traits into the new varieties that make them easy to spot for a field-based weeding robot. For example, the desired variety could express pigments such as anthocyanins or carotenoids in places the wild relative does not, signaling the robot to leave it alone. Alternatively, leaf shape could be changed from serrated to round, or vice versa. The authors point out that these small but meaningful changes in plant phenotype can be made through genome editing, avoiding the pitfalls of introducing foreign genes. (Summary by Mary Williams @PlantTeaching) Trends Plant Sci. 10.1016/j.tplants.2024.03.001
How stems breathe: Oxygen production in woody stems under different light conditions
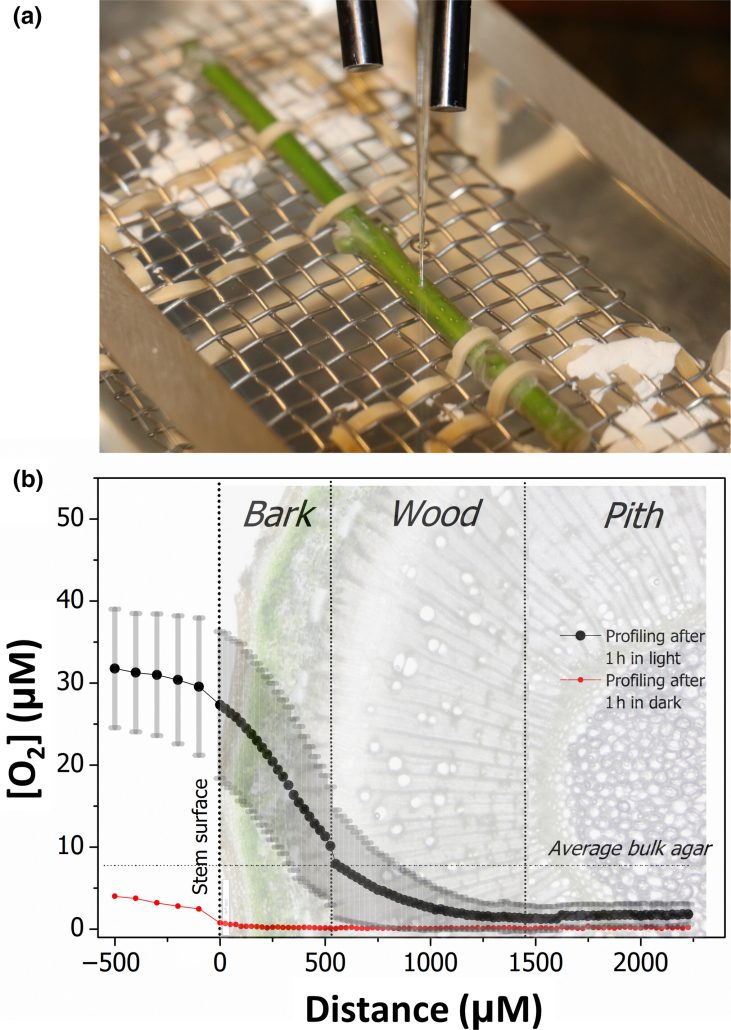 In woody plants, we know much less about how photosynthesis functions in stems than in leaves. This study by Natale et al. focused on how chloroplasts in Fraxinus ornus stems of different ages work. The authors looked at oxygen production rates in whole stems, bark, and wood using various methods like oxygen sensors and fluorescence lifetime imaging microscopy (FLIM). The findings showed that stems produce oxygen when exposed to light, with younger stems producing more oxygen and having higher dark respiration rates than older ones. Bark was the main source of oxygen production in light, but woody parenchyma also contributed as its chloroplasts were found to be active. The authors note that photosynthesis in these deep tissues can be supported by CO2 released from respiration, and the resultant O2 can help prevent hypoxia. FLIM analysis revealed that wood had less photosystem I compared to bark, suggesting that stem chloroplasts adapt to the light quality reaching different parts of the plant. (Summary by Yueh Cho @YuehCho1984) New Phytol. 10.1111/nph.19782
In woody plants, we know much less about how photosynthesis functions in stems than in leaves. This study by Natale et al. focused on how chloroplasts in Fraxinus ornus stems of different ages work. The authors looked at oxygen production rates in whole stems, bark, and wood using various methods like oxygen sensors and fluorescence lifetime imaging microscopy (FLIM). The findings showed that stems produce oxygen when exposed to light, with younger stems producing more oxygen and having higher dark respiration rates than older ones. Bark was the main source of oxygen production in light, but woody parenchyma also contributed as its chloroplasts were found to be active. The authors note that photosynthesis in these deep tissues can be supported by CO2 released from respiration, and the resultant O2 can help prevent hypoxia. FLIM analysis revealed that wood had less photosystem I compared to bark, suggesting that stem chloroplasts adapt to the light quality reaching different parts of the plant. (Summary by Yueh Cho @YuehCho1984) New Phytol. 10.1111/nph.19782
CAM evolution is associated with gene family expansion in an explosive bromeliad radiation
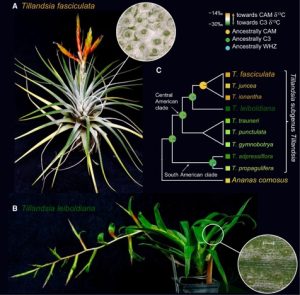 Studies on Crassulacean Acid Metabolism (CAM) plant genomes are scarse and CAM evolution is known to be an ecological driver of diversification. The subgenus Tillandsia (Bromeliaceae) is one of the fastest diversifying clades in the plant kingdom and is known for its adaptive CAM trait. It has recently been subect to comparative genomic studies by Crego et al. CAM evolution is known to be mostly driven by gene family expansion, hence the authors studied two ecologically divergent members of subgenus Tillandsia, Tillandsia fasculata (a CAM plant) and Tillandsia leiboldiana, a C3 plant. De-novo haploid genome assembly of these two species were constructed. Gene family expansion in T. fasciculata was greater than T. leiboldiana in terms of increased number of genes belonging to multicopy families followed by a higher rate of gene duplication. In-depth gene expression analyis on the two genomes also showed differences related to CAM evolution, primarily at the regulatory level. CAM-related genes showed differential expression (DE) between both species across a 24h period. CAM-related DE-gene expression profiles are more aligned to the T. fasciculata genome, possibly leading to its CAM phenotype. Integration of such de novo assembly studies with in-depth transcriptome analysis to understand complex traits like CAM evolution is important in context of evolutionary biology studies on this highly species-rich plant family. (Summary by Indrani Kakati Baruah @Indranik333) Plant Cell 10.1093/plcell/koae130
Studies on Crassulacean Acid Metabolism (CAM) plant genomes are scarse and CAM evolution is known to be an ecological driver of diversification. The subgenus Tillandsia (Bromeliaceae) is one of the fastest diversifying clades in the plant kingdom and is known for its adaptive CAM trait. It has recently been subect to comparative genomic studies by Crego et al. CAM evolution is known to be mostly driven by gene family expansion, hence the authors studied two ecologically divergent members of subgenus Tillandsia, Tillandsia fasculata (a CAM plant) and Tillandsia leiboldiana, a C3 plant. De-novo haploid genome assembly of these two species were constructed. Gene family expansion in T. fasciculata was greater than T. leiboldiana in terms of increased number of genes belonging to multicopy families followed by a higher rate of gene duplication. In-depth gene expression analyis on the two genomes also showed differences related to CAM evolution, primarily at the regulatory level. CAM-related genes showed differential expression (DE) between both species across a 24h period. CAM-related DE-gene expression profiles are more aligned to the T. fasciculata genome, possibly leading to its CAM phenotype. Integration of such de novo assembly studies with in-depth transcriptome analysis to understand complex traits like CAM evolution is important in context of evolutionary biology studies on this highly species-rich plant family. (Summary by Indrani Kakati Baruah @Indranik333) Plant Cell 10.1093/plcell/koae130
Triacylglycerol remodeling to synthesize unusual fatty acids in plants
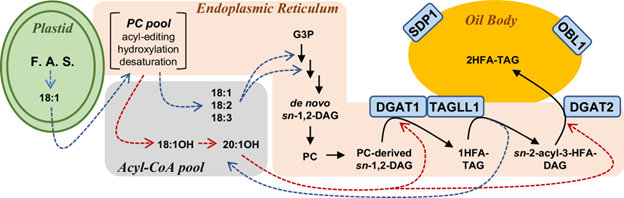 Plant oils, composed of fatty acids, provide humans and animals with food, essential nutrients, chemicals, and biofuels. Plants accumulate over 450 “unusual” fatty acids, which could hold significant value if they achieve optimal production. Hydroxylated fatty acids (HFA) are unusual fatty acids that have applications in industrial production of paints, polymers, and cosmetics. Attempts to engineer crops with HFA have resulted in low HFA levels and decreased seed oil phenotypes due to bottlenecks in overlapping pathways of oil and membrane lipid synthesis, where HFA is incompatible. Parchuri et al. discovered a new mechanism in the brassica Physaria fendleri that naturally overcomes these bottlenecks via remodeling of the initially synthesized triacylglycerol (TAG) with HFA. The TAG remodeling mechanism involves the concerted action of typical fatty acid-specific lipase and HFA-specific acyltransferase enzymes. The authors further engineered this mechanism in Arabidopsis, employing it as a genetic engineering tool to enhance HFA levels in seed oil. In the future, TAG remodeling can be induced in other crops for the production of nutritionally and industrially valuable fatty acids in seed oil. (Summary by Maneesh Lingwan @LingwanManeesh) Nature Comms. 10.1038/s41467-024-47995-x
Plant oils, composed of fatty acids, provide humans and animals with food, essential nutrients, chemicals, and biofuels. Plants accumulate over 450 “unusual” fatty acids, which could hold significant value if they achieve optimal production. Hydroxylated fatty acids (HFA) are unusual fatty acids that have applications in industrial production of paints, polymers, and cosmetics. Attempts to engineer crops with HFA have resulted in low HFA levels and decreased seed oil phenotypes due to bottlenecks in overlapping pathways of oil and membrane lipid synthesis, where HFA is incompatible. Parchuri et al. discovered a new mechanism in the brassica Physaria fendleri that naturally overcomes these bottlenecks via remodeling of the initially synthesized triacylglycerol (TAG) with HFA. The TAG remodeling mechanism involves the concerted action of typical fatty acid-specific lipase and HFA-specific acyltransferase enzymes. The authors further engineered this mechanism in Arabidopsis, employing it as a genetic engineering tool to enhance HFA levels in seed oil. In the future, TAG remodeling can be induced in other crops for the production of nutritionally and industrially valuable fatty acids in seed oil. (Summary by Maneesh Lingwan @LingwanManeesh) Nature Comms. 10.1038/s41467-024-47995-x
A novel BZR/BES transcription factor controls the development of haploid reproductive organs in Marchantia polymorpha
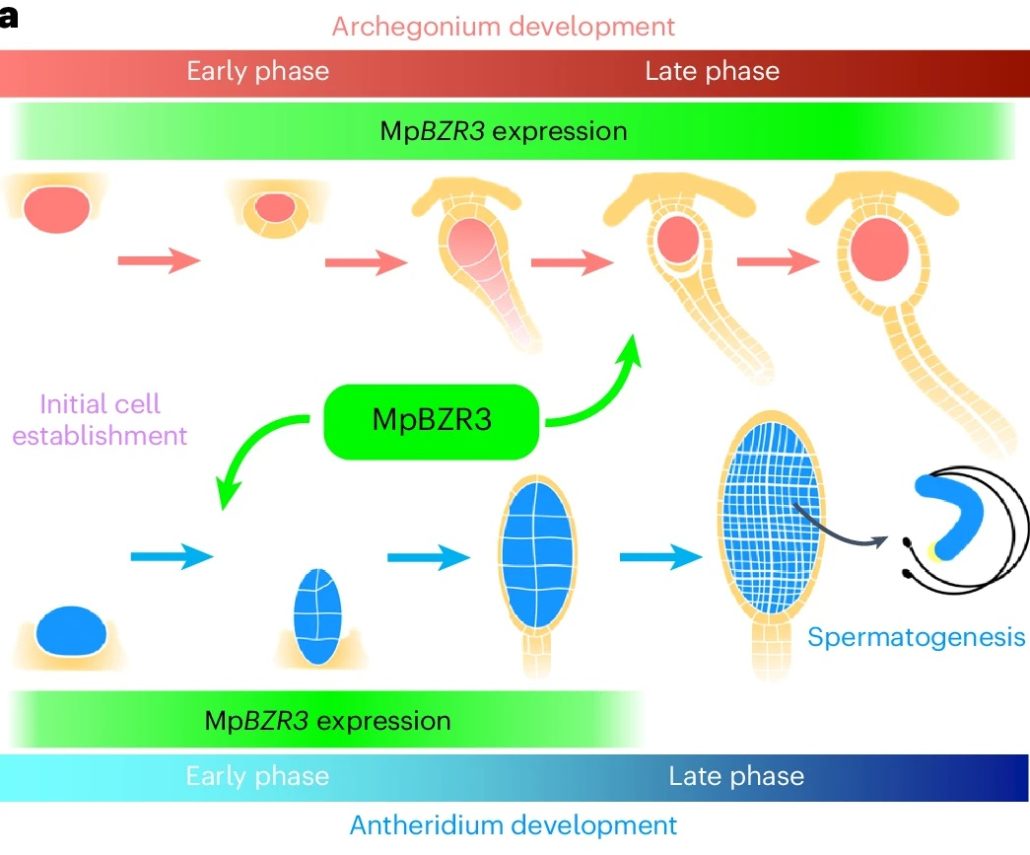 Gametogenesis is essential for sexual reproduction. In bryophytes, lycophytes, and ferns, gametogenesis takes place in gametangia: antheridia for sperm production and archegonia for egg production. How these specialized reproductive organs develop at the molecular level remains unclear. Furuya et al. identified a unique BZR/BES transcription factor, MpBZR3, that regulates gametangium development in Marchantia polymorpha. Overexpression of MpBZR3 caused unusual gametangia growth. MpBZR3 is essential for the proper development of both types of gametangia and acts differently in male and female plants. In male plants it supports early antheridia development and in female plants is supports later archegonia development. This specialization of MpBZR3 may have contributed to the evolution of plant reproductive systems. (Summary by Yueh Cho @YuehCho1984) Nature Plant. 10.1038/s41477-024-01669-0
Gametogenesis is essential for sexual reproduction. In bryophytes, lycophytes, and ferns, gametogenesis takes place in gametangia: antheridia for sperm production and archegonia for egg production. How these specialized reproductive organs develop at the molecular level remains unclear. Furuya et al. identified a unique BZR/BES transcription factor, MpBZR3, that regulates gametangium development in Marchantia polymorpha. Overexpression of MpBZR3 caused unusual gametangia growth. MpBZR3 is essential for the proper development of both types of gametangia and acts differently in male and female plants. In male plants it supports early antheridia development and in female plants is supports later archegonia development. This specialization of MpBZR3 may have contributed to the evolution of plant reproductive systems. (Summary by Yueh Cho @YuehCho1984) Nature Plant. 10.1038/s41477-024-01669-0
Transcriptional changes during barley grain development
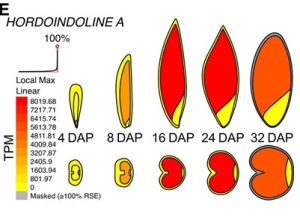 Barley is a globally important cereal crop, so understanding barley grain development is of much interest. Here, Kovacik et al. investigated transcriptional changes in barley grains at five points across development, 4, 8, 16, 24 and 32 days after pollination. For each time point, grains were manually dissected into embryo, endosperm and seed maternal tissues, flow cytometry was used to check the purity of the dissections, and samples were sent for RNA sequencing. Principal component analysis revealed that there was large transcriptional reprogramming between 8 and 24 days after pollination, whilst there were fewer transcriptional changes at later stages of development (24-32 days after pollination). GO term analysis showed that pathways were differentially expressed in different tissues, for example isoprenoid biosynthesis genes were specifically upregulated in seed maternal tissues between 4 and 16 days after pollination, whilst mRNA splicing genes were specifically enriched in the embryo at 16 and 24 days after pollination. All in all, this paper contains a plethora of transcriptional data which enhances our understanding of barley grain development and notably, the data has been uploaded onto BAR ePlant so it is easily accessible. (Summary by Rose McNelly @Rose_McN) Plant Cell 10.1093/plcell/koae095
Barley is a globally important cereal crop, so understanding barley grain development is of much interest. Here, Kovacik et al. investigated transcriptional changes in barley grains at five points across development, 4, 8, 16, 24 and 32 days after pollination. For each time point, grains were manually dissected into embryo, endosperm and seed maternal tissues, flow cytometry was used to check the purity of the dissections, and samples were sent for RNA sequencing. Principal component analysis revealed that there was large transcriptional reprogramming between 8 and 24 days after pollination, whilst there were fewer transcriptional changes at later stages of development (24-32 days after pollination). GO term analysis showed that pathways were differentially expressed in different tissues, for example isoprenoid biosynthesis genes were specifically upregulated in seed maternal tissues between 4 and 16 days after pollination, whilst mRNA splicing genes were specifically enriched in the embryo at 16 and 24 days after pollination. All in all, this paper contains a plethora of transcriptional data which enhances our understanding of barley grain development and notably, the data has been uploaded onto BAR ePlant so it is easily accessible. (Summary by Rose McNelly @Rose_McN) Plant Cell 10.1093/plcell/koae095
Antibody array-based proteome approach reveals proteins involved in grape seed development
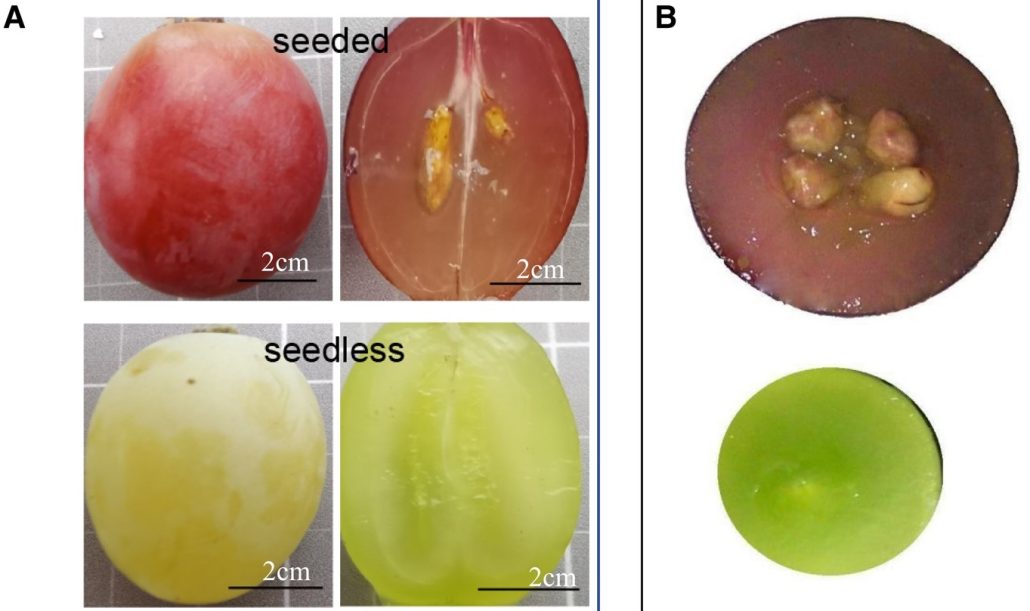 Grape (Vitis vinifera) is a globally cultivated fruit for fresh or processed consumption, and seedless grapes are highly preferred for consumer convenience. In seedless grapes, fertilization occurs but the embryo subsequently aborts, although the molecular basis for this abortion is not understood. Zhang et al. used a special antibody array-based proteome approach to identify differences between seeded and seedless grapes. Validation and mass spectrometry tests confirmed the 71 differentially-regulated proteins. A protein-protein interaction network showed changes in pathways related to carbon metabolism and glycolysis. When the authors overexpressed a protein called VvDUF642 in tomato plants, fewer seeds were produced. VvDUF642 works with another protein, VvPAE, which also reduces seed production when overexpressed. Thus, this study identified proteins that are involved in seed formation and these insights could help breed seedless grapes and other fruits in the future. (Summary by Yueh Cho @YuehCho1984) Plant Physiol. 10.1093/plphys/kiad682
Grape (Vitis vinifera) is a globally cultivated fruit for fresh or processed consumption, and seedless grapes are highly preferred for consumer convenience. In seedless grapes, fertilization occurs but the embryo subsequently aborts, although the molecular basis for this abortion is not understood. Zhang et al. used a special antibody array-based proteome approach to identify differences between seeded and seedless grapes. Validation and mass spectrometry tests confirmed the 71 differentially-regulated proteins. A protein-protein interaction network showed changes in pathways related to carbon metabolism and glycolysis. When the authors overexpressed a protein called VvDUF642 in tomato plants, fewer seeds were produced. VvDUF642 works with another protein, VvPAE, which also reduces seed production when overexpressed. Thus, this study identified proteins that are involved in seed formation and these insights could help breed seedless grapes and other fruits in the future. (Summary by Yueh Cho @YuehCho1984) Plant Physiol. 10.1093/plphys/kiad682
Gravitropism with a pinch of salt: Changes in cell wall composition modulate root growth direction in saline conditions
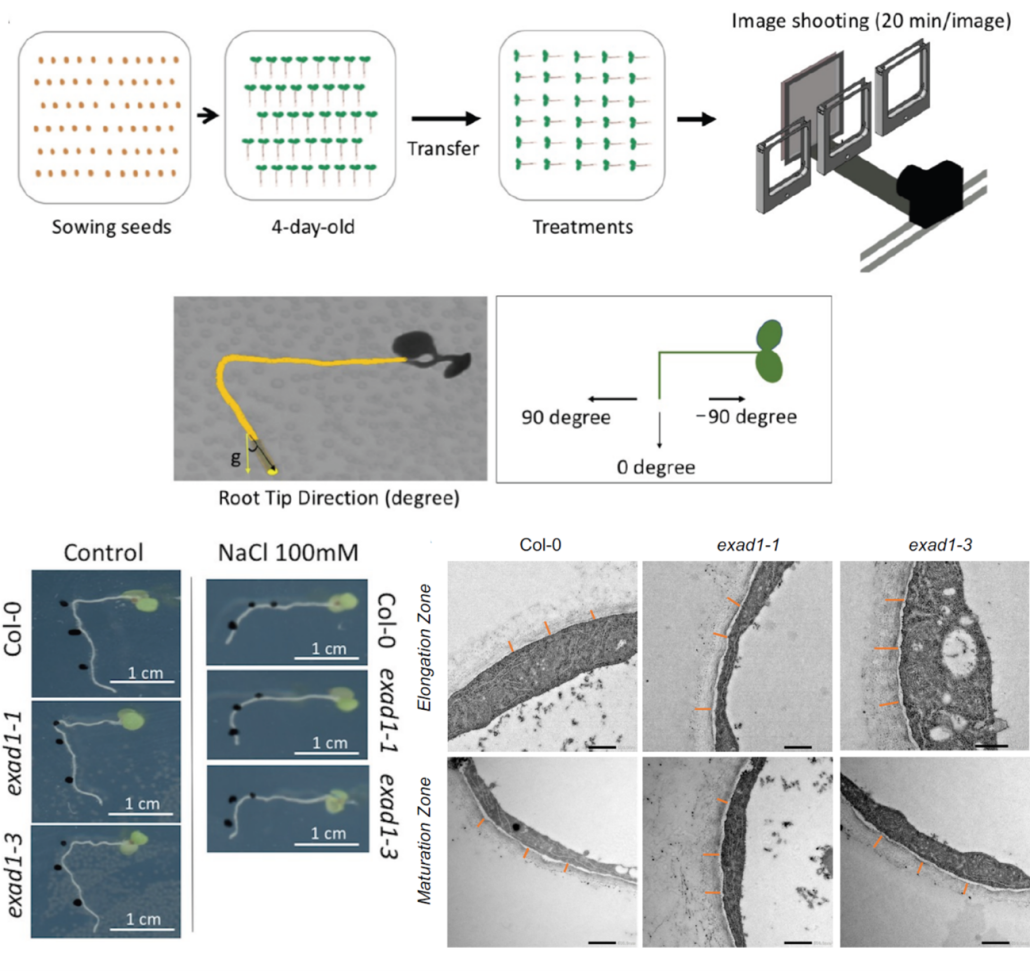 Soil salinization causes massive yield losses in agriculture, and its impact on plants goes beyond what our eyes can see. Roots are immediately affected by the direct exposure to a salt-(NaCl) rich substrate. Here, Zou et al. investigate the salt-induced altered root gravitropic responses in Arabidopsis thaliana, dissecting the role of cell wall modification in this process. The authors developed a phenotyping tool termed Salt-Induced Tilting Assay, which allowed them to reveal that altered salt-induced gravitropic responses are linked to sodium ions, not to osmotic stress. In search for the genetic elements behind this phenotype, a genome-wide association study of 345 Arabidopsis accessions underlined a potential role for EXTENSIN ARABINOSE DEFICIENT TRANSFERASE (ExAD). Extensins, key cell wall glycoproteins, are modified by ExAD through the addition of an arabinose residue (arabinosylation). Root direction angles of loss-of-function mutants exad1-1 and exad1-3 differed from those of Col-0 under saline conditions, as did arabinose levels, which were mildly lower due to extensins lacking ExAD-mediated arabinosylation. Mutants exad1-1 and exad1-3 had thicker, more porous, and less dense cell walls than Col-0, further illustrating the importance of arabinosylated extensins in preserving cell wall integrity under saline conditions. (Summary by John Vilasboa @vilasjohn) Plant Cell 10.1093/plcell/koae135
Soil salinization causes massive yield losses in agriculture, and its impact on plants goes beyond what our eyes can see. Roots are immediately affected by the direct exposure to a salt-(NaCl) rich substrate. Here, Zou et al. investigate the salt-induced altered root gravitropic responses in Arabidopsis thaliana, dissecting the role of cell wall modification in this process. The authors developed a phenotyping tool termed Salt-Induced Tilting Assay, which allowed them to reveal that altered salt-induced gravitropic responses are linked to sodium ions, not to osmotic stress. In search for the genetic elements behind this phenotype, a genome-wide association study of 345 Arabidopsis accessions underlined a potential role for EXTENSIN ARABINOSE DEFICIENT TRANSFERASE (ExAD). Extensins, key cell wall glycoproteins, are modified by ExAD through the addition of an arabinose residue (arabinosylation). Root direction angles of loss-of-function mutants exad1-1 and exad1-3 differed from those of Col-0 under saline conditions, as did arabinose levels, which were mildly lower due to extensins lacking ExAD-mediated arabinosylation. Mutants exad1-1 and exad1-3 had thicker, more porous, and less dense cell walls than Col-0, further illustrating the importance of arabinosylated extensins in preserving cell wall integrity under saline conditions. (Summary by John Vilasboa @vilasjohn) Plant Cell 10.1093/plcell/koae135
Volatile communication in plants relies on a KAI2-mediated signaling pathway
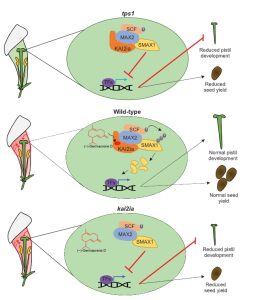 While it is recognized that plant communication, both within and between plants, can be achieved through emission and perception of volatile organic compounds (VOCs), it is unclear exactly how these signals are perceived and transmitted. Stirling et al. tease apart the components of within-plant VOC communication using the clever model system of pistil development in Petunia hybrida and a set of genetic and biochemical experiments. The authors show that changes in pistil width and length are dependent on emission of the VOC (-)-germacrene-D from the flower tube, and that this signal is perceived by the receptor karrikin insensitive2ia (KAI2ia), so that defects in either of these components prevent normal pistil development. KAI2ia is a homologue of karrikin insensitive2 (KIA2), previously identified as a receptor for volatile karrikins produced during fires. KIA2 interacts with MAX1 to degrade transcriptional repressors and activate fire-related responses. The authors show that KAI2ia acts through a similar mechanism. Many plants synthesise (-)-germacrene-D and receptors for this VOC are found in some insect pollinators, suggesting KAI2-like receptors may also have other roles such as in plant-insect communication. (Summary by Alicia Quinn @AliciaQuinnSci) Science 10.1126/science.adl4685
While it is recognized that plant communication, both within and between plants, can be achieved through emission and perception of volatile organic compounds (VOCs), it is unclear exactly how these signals are perceived and transmitted. Stirling et al. tease apart the components of within-plant VOC communication using the clever model system of pistil development in Petunia hybrida and a set of genetic and biochemical experiments. The authors show that changes in pistil width and length are dependent on emission of the VOC (-)-germacrene-D from the flower tube, and that this signal is perceived by the receptor karrikin insensitive2ia (KAI2ia), so that defects in either of these components prevent normal pistil development. KAI2ia is a homologue of karrikin insensitive2 (KIA2), previously identified as a receptor for volatile karrikins produced during fires. KIA2 interacts with MAX1 to degrade transcriptional repressors and activate fire-related responses. The authors show that KAI2ia acts through a similar mechanism. Many plants synthesise (-)-germacrene-D and receptors for this VOC are found in some insect pollinators, suggesting KAI2-like receptors may also have other roles such as in plant-insect communication. (Summary by Alicia Quinn @AliciaQuinnSci) Science 10.1126/science.adl4685
Spatial co-transcriptomics reveals discrete stages of the arbuscular mycorrhizal symbiosis
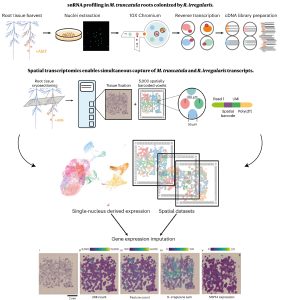 In recent years, single-cell RNA sequencing (scRNA-seq) and single-nuclei RNA sequencing (snRNA-seq) techniques have revolutionized plant biology by enabling the identification of novel cell types, modeling developmental trajectories, and analyzing transcriptional activity at the cellular level. However, these methods lack spatial context upon cell dissociation from tissues. Spatial transcriptomics (ST) overcomes this, preserving positional data and offering profound insights into cellular interactions. Serrano et. al. created the first high-resolution spatially resolved integrated map of a multi-kingdom symbiotic interaction by analyzing the transcriptomes the model legume Medicago truncatula and the fungus Rhizophagus irregularis in arbuscular mycorrhizal (AM) symbiosis at cellular and spatial resolution. Their observations revealed dynamic changes in the transcriptome profiles of M. truncatula cortex cells during different stages of colonization by R. irregularis, highlighting the dynamic interplay between both organisms during symbiosis establishment. They identified a spatial cluster as AM-responsive based on the expression of marker genes for the symbiosis from both organisms. The transcripts within this cluster, along with R. irregularis genes expressed within the mycorrhizal capture areas, denote promising targets for functional characterization studies in both partners, offering potential benefits to the AM community in uncovering new functionalities. The generated transcriptomic map highlights the value of novel multi-omics approaches in answering biological questions and facilitating targeted interventions in agricultural practices and ecosystem management. (Summary by Ileana Tossolini @IleanaDrt) Nature Plants 10.1038/s41477-024-01666-3.
In recent years, single-cell RNA sequencing (scRNA-seq) and single-nuclei RNA sequencing (snRNA-seq) techniques have revolutionized plant biology by enabling the identification of novel cell types, modeling developmental trajectories, and analyzing transcriptional activity at the cellular level. However, these methods lack spatial context upon cell dissociation from tissues. Spatial transcriptomics (ST) overcomes this, preserving positional data and offering profound insights into cellular interactions. Serrano et. al. created the first high-resolution spatially resolved integrated map of a multi-kingdom symbiotic interaction by analyzing the transcriptomes the model legume Medicago truncatula and the fungus Rhizophagus irregularis in arbuscular mycorrhizal (AM) symbiosis at cellular and spatial resolution. Their observations revealed dynamic changes in the transcriptome profiles of M. truncatula cortex cells during different stages of colonization by R. irregularis, highlighting the dynamic interplay between both organisms during symbiosis establishment. They identified a spatial cluster as AM-responsive based on the expression of marker genes for the symbiosis from both organisms. The transcripts within this cluster, along with R. irregularis genes expressed within the mycorrhizal capture areas, denote promising targets for functional characterization studies in both partners, offering potential benefits to the AM community in uncovering new functionalities. The generated transcriptomic map highlights the value of novel multi-omics approaches in answering biological questions and facilitating targeted interventions in agricultural practices and ecosystem management. (Summary by Ileana Tossolini @IleanaDrt) Nature Plants 10.1038/s41477-024-01666-3.
Development of a low-cost plant phenotyping facility
 New technologies, like personal computers or smart phones, often have limited adoption due to their high cost or requirement for advanced technological skills. Greater affordability and ease of use leads to greater adoption. Here, Yu, Sussman et al. describe the development of an affordable, portable plant phenotyping platform and pipeline that is easy to build and use. For the image collection station, they employ low-cost Raspberry Pi cameras and computers, wooden frames, LED lights, and 3D-printed components to hold everything in place. To this they add a workflow, RaspiPheno Pipe and RaspiPheno App (https://github.com/Leon-Yu0320/BTI-Plant-phenotyping/tree/main/RasPiPheno_APP) that automates image analysis and makes these steps accessible even to those without extensive computer skills. As proof of concept, they used this platform for a phenotypic analysis of stress responses using Arabidopsis, cowpea (Vigna unguiculata), and tepary bean (Phaseolus acutifolius). Through imaging of a cowpea diversity panel and GWAS, they identified several loci associated with drought resilience, and tested them functionally through analysis of Arabidopsis T-DNA mutants. The manuscript includes extensive notes on building and using these low-cost phenotyping facilities, supplemented by additional information at protocols.io. (Summary by Mary Williams @PlantTeaching) Plant Physiol. 10.1093/plphys/kiae237
New technologies, like personal computers or smart phones, often have limited adoption due to their high cost or requirement for advanced technological skills. Greater affordability and ease of use leads to greater adoption. Here, Yu, Sussman et al. describe the development of an affordable, portable plant phenotyping platform and pipeline that is easy to build and use. For the image collection station, they employ low-cost Raspberry Pi cameras and computers, wooden frames, LED lights, and 3D-printed components to hold everything in place. To this they add a workflow, RaspiPheno Pipe and RaspiPheno App (https://github.com/Leon-Yu0320/BTI-Plant-phenotyping/tree/main/RasPiPheno_APP) that automates image analysis and makes these steps accessible even to those without extensive computer skills. As proof of concept, they used this platform for a phenotypic analysis of stress responses using Arabidopsis, cowpea (Vigna unguiculata), and tepary bean (Phaseolus acutifolius). Through imaging of a cowpea diversity panel and GWAS, they identified several loci associated with drought resilience, and tested them functionally through analysis of Arabidopsis T-DNA mutants. The manuscript includes extensive notes on building and using these low-cost phenotyping facilities, supplemented by additional information at protocols.io. (Summary by Mary Williams @PlantTeaching) Plant Physiol. 10.1093/plphys/kiae237


 Finding: We report on the ability of some maize genotypes to alter the azimuthal orientations of their leaves during development in coordination with adjacent plants. Although the upper canopies of these genotypes retain the typical alternate-distichous phyllotaxy of maize, their leaves grow parallel to those of adjacent plants. A genome-wide association study conducted on this parallel canopy trait and on the fraction of intercepted photosynthetically active radiation identified candidate genes, many of which are associated with shade avoidance syndrome and ligule development, such as liguleless1 (lg1). Further, mutagenesis studies demonstrated liguleless gene functions are required for normal light responses, including azimuthal canopy re-orientation.
Finding: We report on the ability of some maize genotypes to alter the azimuthal orientations of their leaves during development in coordination with adjacent plants. Although the upper canopies of these genotypes retain the typical alternate-distichous phyllotaxy of maize, their leaves grow parallel to those of adjacent plants. A genome-wide association study conducted on this parallel canopy trait and on the fraction of intercepted photosynthetically active radiation identified candidate genes, many of which are associated with shade avoidance syndrome and ligule development, such as liguleless1 (lg1). Further, mutagenesis studies demonstrated liguleless gene functions are required for normal light responses, including azimuthal canopy re-orientation. Findings: Using a comprehensive approach that includes cell biology, X-ray crystallography, biochemistry, electron microscopy, and plant physiology, our study reveals CHUP1 as a type of actin nucleator controlling chloroplast movement. Noteworthy discoveries include CHUP1’s pivotal role in polymerizing cp-actin filaments under weak light and depolymerizing them under strong light, in a blue light- and phototropin-dependent manner. Notably, the C-terminal half of CHUP1 (CHUP1-C) exhibits structural similarity to the formin FH-2 domain, despite lacking amino acid sequence homology. Furthermore, CHUP1-C’s ability to promote actin polymerization when complexed with profilin in vitro implies that CHUP1 evolved independently as a plant-specific actin nucleator for chloroplast movement.
Findings: Using a comprehensive approach that includes cell biology, X-ray crystallography, biochemistry, electron microscopy, and plant physiology, our study reveals CHUP1 as a type of actin nucleator controlling chloroplast movement. Noteworthy discoveries include CHUP1’s pivotal role in polymerizing cp-actin filaments under weak light and depolymerizing them under strong light, in a blue light- and phototropin-dependent manner. Notably, the C-terminal half of CHUP1 (CHUP1-C) exhibits structural similarity to the formin FH-2 domain, despite lacking amino acid sequence homology. Furthermore, CHUP1-C’s ability to promote actin polymerization when complexed with profilin in vitro implies that CHUP1 evolved independently as a plant-specific actin nucleator for chloroplast movement. Next steps: We are working on elucidating the molecular mechanism underlying MORC6-mediated selective rRNA gene silencing. We will determine DNA methylation levels of 45S rRNA genes and detect NOR structures in morc6 mutants.
Next steps: We are working on elucidating the molecular mechanism underlying MORC6-mediated selective rRNA gene silencing. We will determine DNA methylation levels of 45S rRNA genes and detect NOR structures in morc6 mutants.