Spatial co-transcriptomics reveals discrete stages of the arbuscular mycorrhizal symbiosis
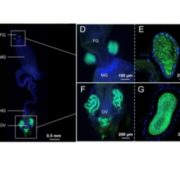
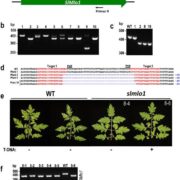
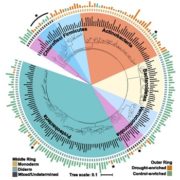
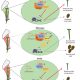
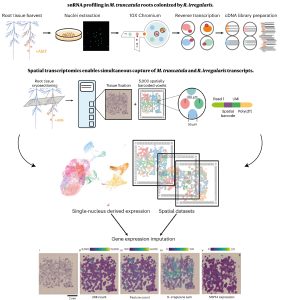 In recent years, single-cell RNA sequencing (scRNA-seq) and single-nuclei RNA sequencing (snRNA-seq) techniques have revolutionized plant biology by enabling the identification of novel cell types, modeling developmental trajectories, and analyzing transcriptional activity at the cellular level. However, these methods lack spatial context upon cell dissociation from tissues. Spatial transcriptomics (ST) overcomes this, preserving positional data and offering profound insights into cellular interactions. Serrano et. al. created the first high-resolution spatially resolved integrated map of a multi-kingdom symbiotic interaction by analyzing the transcriptomes the model legume Medicago truncatula and the fungus Rhizophagus irregularis in arbuscular mycorrhizal (AM) symbiosis at cellular and spatial resolution. Their observations revealed dynamic changes in the transcriptome profiles of M. truncatula cortex cells during different stages of colonization by R. irregularis, highlighting the dynamic interplay between both organisms during symbiosis establishment. They identified a spatial cluster as AM-responsive based on the expression of marker genes for the symbiosis from both organisms. The transcripts within this cluster, along with R. irregularis genes expressed within the mycorrhizal capture areas, denote promising targets for functional characterization studies in both partners, offering potential benefits to the AM community in uncovering new functionalities. The generated transcriptomic map highlights the value of novel multi-omics approaches in answering biological questions and facilitating targeted interventions in agricultural practices and ecosystem management. (Summary by Ileana Tossolini @IleanaDrt) Nature Plants 10.1038/s41477-024-01666-3.
In recent years, single-cell RNA sequencing (scRNA-seq) and single-nuclei RNA sequencing (snRNA-seq) techniques have revolutionized plant biology by enabling the identification of novel cell types, modeling developmental trajectories, and analyzing transcriptional activity at the cellular level. However, these methods lack spatial context upon cell dissociation from tissues. Spatial transcriptomics (ST) overcomes this, preserving positional data and offering profound insights into cellular interactions. Serrano et. al. created the first high-resolution spatially resolved integrated map of a multi-kingdom symbiotic interaction by analyzing the transcriptomes the model legume Medicago truncatula and the fungus Rhizophagus irregularis in arbuscular mycorrhizal (AM) symbiosis at cellular and spatial resolution. Their observations revealed dynamic changes in the transcriptome profiles of M. truncatula cortex cells during different stages of colonization by R. irregularis, highlighting the dynamic interplay between both organisms during symbiosis establishment. They identified a spatial cluster as AM-responsive based on the expression of marker genes for the symbiosis from both organisms. The transcripts within this cluster, along with R. irregularis genes expressed within the mycorrhizal capture areas, denote promising targets for functional characterization studies in both partners, offering potential benefits to the AM community in uncovering new functionalities. The generated transcriptomic map highlights the value of novel multi-omics approaches in answering biological questions and facilitating targeted interventions in agricultural practices and ecosystem management. (Summary by Ileana Tossolini @IleanaDrt) Nature Plants 10.1038/s41477-024-01666-3.