No-Genome-Required-GWAS (Nature Genetics)
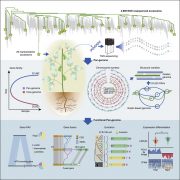
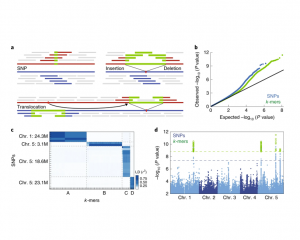 Conventional approaches to connect phenotype to genotype, including genome-wide association studies (GWAS), are often limited by the quality of the species’ reference genome, and frequently neglect to detect structural variants that are common in plant genomes. Here, Voichek and Weigel present a “No-Genome-Required-GWAS” approach (summarized in this Twitter thread), which is essentially the reverse of a conventional GWAS, that avoids using a reference genome and single nucleotide polymorphism (SNP) data. Instead, short sequences called k-mers (derived from raw sequencing data) are associated with a phenotype, and only thereafter the sequence’s genomic context is uncovered. As a proof of principle, the authors conducted a k-mer-based GWAS for flowering time in Arabidopsis, that identified associations previously detected by SNPs. The authors re-analyzed 2,000 traits in Arabidopsis, maize, and tomato, with case studies demonstrating the advantages of the k-mer-based approach versus that of SNPs. In all species, top k-mer associations were typically stronger than top SNPs. The k-mer-based approach also revealed new associations with structural variants and with regions absent from reference genomes. Moreover, k-mers successfully estimated kinship between individuals, meaning this method can be applied to species lacking high-quality reference genomes. This study emphasizes the merits of a reference genome-independent approach to detect new genetic variants, even in thoroughly studied plant species. The No-Genome-Required-GWAS has exciting relevance for linking phenotype to genotype in both model and non-model species alike. (Summary by Caroline Dowling @CarolineD0wling) Nature Genetics 10.1038/s41588-020-0612-7
Conventional approaches to connect phenotype to genotype, including genome-wide association studies (GWAS), are often limited by the quality of the species’ reference genome, and frequently neglect to detect structural variants that are common in plant genomes. Here, Voichek and Weigel present a “No-Genome-Required-GWAS” approach (summarized in this Twitter thread), which is essentially the reverse of a conventional GWAS, that avoids using a reference genome and single nucleotide polymorphism (SNP) data. Instead, short sequences called k-mers (derived from raw sequencing data) are associated with a phenotype, and only thereafter the sequence’s genomic context is uncovered. As a proof of principle, the authors conducted a k-mer-based GWAS for flowering time in Arabidopsis, that identified associations previously detected by SNPs. The authors re-analyzed 2,000 traits in Arabidopsis, maize, and tomato, with case studies demonstrating the advantages of the k-mer-based approach versus that of SNPs. In all species, top k-mer associations were typically stronger than top SNPs. The k-mer-based approach also revealed new associations with structural variants and with regions absent from reference genomes. Moreover, k-mers successfully estimated kinship between individuals, meaning this method can be applied to species lacking high-quality reference genomes. This study emphasizes the merits of a reference genome-independent approach to detect new genetic variants, even in thoroughly studied plant species. The No-Genome-Required-GWAS has exciting relevance for linking phenotype to genotype in both model and non-model species alike. (Summary by Caroline Dowling @CarolineD0wling) Nature Genetics 10.1038/s41588-020-0612-7
[altmetric doi=”10.1038/s41588-020-0612-7″ details=”right” float=”right”]