Pan-genome of wild and cultivated soybeans (Cell)
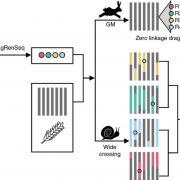
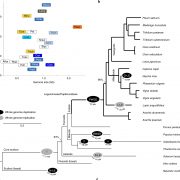
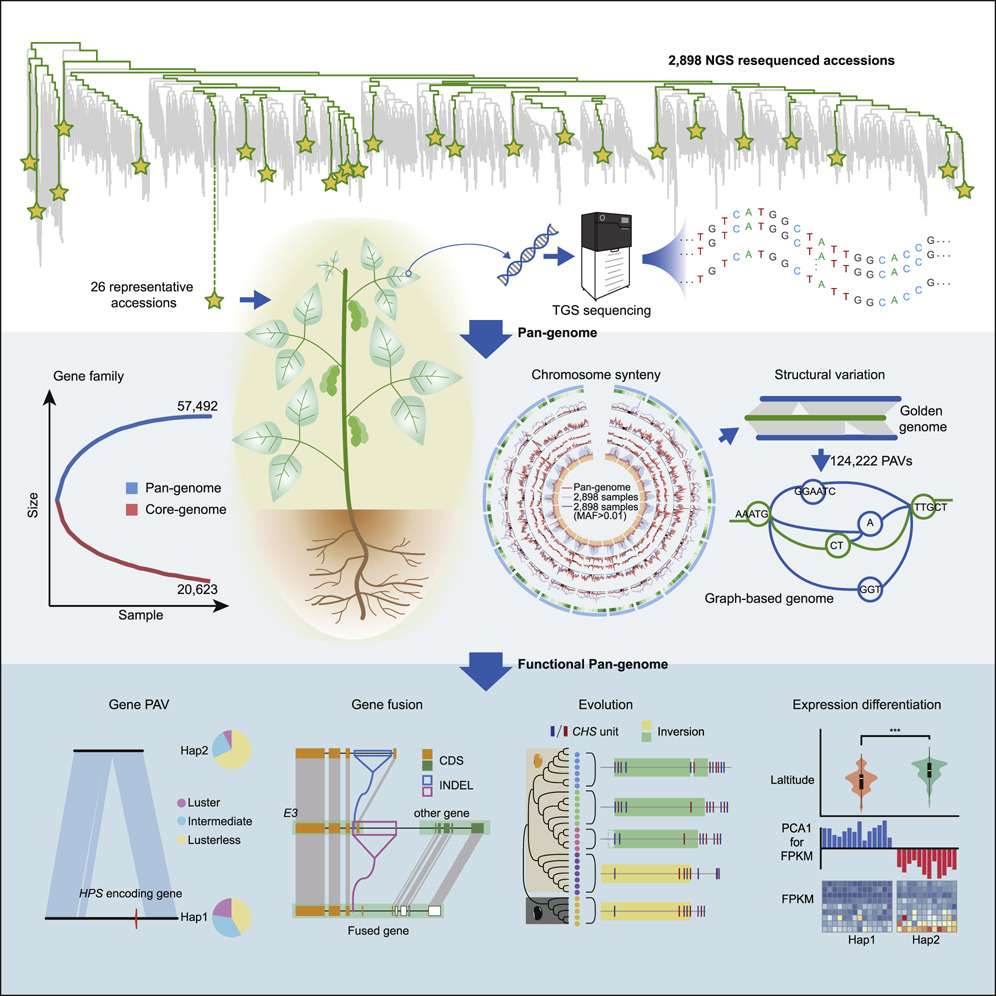 Widespread genome sequencing of individuals has revealed the high level of intraspecific variability in plant species. As such, constructing high-quality pan-genomes is a growing necessity to study dynamic plant genomes. Here, Liu et al. release the soybean pan-genome formed from sequencing 2,898 accessions, including landraces, wild, and cultivated soybeans. The authors de novo assembled 26 accessions which were carefully selected to represent the geographical distributions and phylogenetics of the 2,898 accessions. A graph-based genome was subsequently formed, onto which short reads of the 2,898 accessions were mapped, identifying over 55,000 structural variants (SVs). The authors found 15 gene fusion events in different accessions, including a gene fusion involving E3, a PHYA ortholog. Furthermore, the contribution of SVs in soybean domestication was highlighted, such as the alteration of seed coat pigmentation. Finally, over 1,000 SVs in core genes (present in all newly assembled accessions) were associated with altered expression levels, and an iron deficiency chlorosis QTL was clarified, with SVs at this locus uncovered in various accessions. The de novo genomes assembled in this study are promising candidates for sophisticated soybean mapping population construction given that they were carefully selected to encompass both phylogenetic and geographic distributions. The publication of this high-quality pan-genome signifies a comprehensive resource that will certainly expedite both future breeding efforts and evolutionary studies in soybean. Summary by Caroline Dowling @CarolineD0wling Cell 10.1016/j.cell.2020.05.023
Widespread genome sequencing of individuals has revealed the high level of intraspecific variability in plant species. As such, constructing high-quality pan-genomes is a growing necessity to study dynamic plant genomes. Here, Liu et al. release the soybean pan-genome formed from sequencing 2,898 accessions, including landraces, wild, and cultivated soybeans. The authors de novo assembled 26 accessions which were carefully selected to represent the geographical distributions and phylogenetics of the 2,898 accessions. A graph-based genome was subsequently formed, onto which short reads of the 2,898 accessions were mapped, identifying over 55,000 structural variants (SVs). The authors found 15 gene fusion events in different accessions, including a gene fusion involving E3, a PHYA ortholog. Furthermore, the contribution of SVs in soybean domestication was highlighted, such as the alteration of seed coat pigmentation. Finally, over 1,000 SVs in core genes (present in all newly assembled accessions) were associated with altered expression levels, and an iron deficiency chlorosis QTL was clarified, with SVs at this locus uncovered in various accessions. The de novo genomes assembled in this study are promising candidates for sophisticated soybean mapping population construction given that they were carefully selected to encompass both phylogenetic and geographic distributions. The publication of this high-quality pan-genome signifies a comprehensive resource that will certainly expedite both future breeding efforts and evolutionary studies in soybean. Summary by Caroline Dowling @CarolineD0wling Cell 10.1016/j.cell.2020.05.023