MtSSPdb—a new data base for the small secreted peptide research community
Eva Hellmann, The Sainsbury Laboratory, University of Cambridge, CB2 1LR Cambridge, United Kingdom, https://orcid.org/0000-0002-4591-2973
Small secreted peptides (SSPs) are short peptides that function as messengers and regulate a variety of processes in plants (Lease and Walker, 2006). They are coded in small open reading frames that give rise to pre-proteins of 100–250 amino acids which then undergo processing into active peptides of 5–50 amino acids. Their small size makes the detection of their open reading frame, expression and peptide itself challenging. SSPs have been shown to regulate plant growth and development and biotic, as well as abiotic interactions between the plant and its environment (Lease and Walker, 2006; Czyzewicz et al., 2013; Matsubayashi, 2014; Breiden and Simon, 2016; De Bang et al., 2017). They are involved in the regulation of nodulation that enables legumes to form symbiotic associations with rhizobia, that can fix nitrogen from the atmosphere and make it available for the plant (Djordjevic et al., 2015). Many crop plants are dependent on synthetic fertiliser, so understanding, and possibly modulating, SSPs is of great interest for the research community and eventually for agriculture.
In their recent publication, Boschiero, Dai and colleagues (2020) present their new SSP database for Medicago: MtSSPdb. This database follows the genome-wide identification and analysis of Medicago SSPs by the same group in 2017 (De Bang et al., 2017).
The authors scanned and re-annotated the M. truncatula genome with special focus on the often overlooked SSP open reading frames. Using Hidden Markov Models (HMMs) of known SSP families for sequence alignments, they identified known and potential new SSP families. A BLAST tool helps to predict the presence of SSPs in a given sequence. Boschiero, Dai and colleagues also compiled available expression data to create a gene expression atlas for M. truncatula SSPs. Another section of the database collects Medicago phenotypes after application of predicted and subsequently synthesised SSPs. While MtSSPdb is currently focused on Medicago, the SSP families previously identified by the same group in maize (Zea mays), tobacco (Nicotiana tabacum) and Arabidopsis (Arabidopsis thaliana) have been included as well (De Bang et al., 2017).
To enable access to their database, Boschiero, Dai and colleagues created a user interface accessible at https://mtsspdb.noble.org/.
Searching the database by entering the gene name, ID, keywords, or the SSP family name will result in a gene card with information on location, sequence, protein properties, SSP family and alignments or SSP family HMM profiles.
It is also possible to BLAST a sequence against the database and analyse it for the presence of potential SSPs. The analysis uses the protein length, the presence of a predicted signal peptide cleavage site, the similarity of HMM patterns or homology to known SSP families and the absence of transmembrane helices to estimate if a SSP is encoded.
In the Gene Expression Atlas, users can search expression patterns of SSPs by keyword, ID or a specific expression pattern in the different RNA-Seq experiments included. Users can perform Co-expression searches as well as GO-term and KEGG-pathway enrichment analyses to learn more about their SSP of interest.
A really exciting feature is the synthetic SSP database where users can see which (hypothetical) SSPs have been synthesised and applied to Medicago to analyse their biological relevance. Aliquots of those synthesised peptides can be requested from the authors.
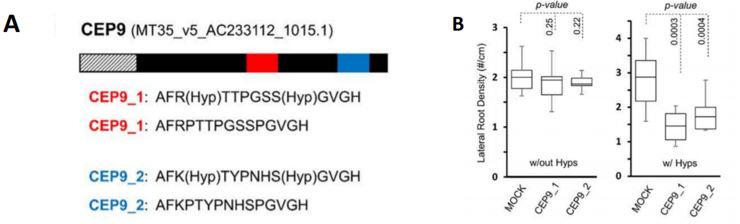
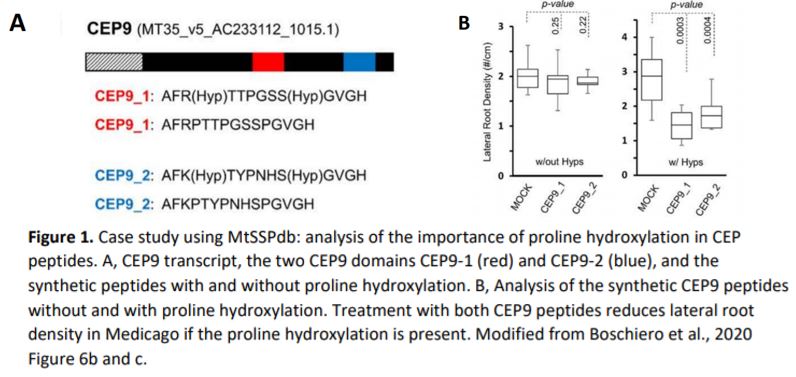 Boschiero, Dai and colleagues provide several case studies for the use of the database. For the example of CEP9 (C-Terminally Encoded Peptide 9), they show how the database helps to identify SSP family members and sequence similarity. CEPs are a highly similar SSP family and CEP1 had been previously shown to inhibit M. truncatula lateral root formation (Imin et al., 2013). The authors found that the CEP9 sequence is very similar to that of CEP1 and predicted that the same residues would require hydroxylation to render the peptide functional. They synthesised CEP1 and CEP9 with and without proline hydroxylation and analysed the effect on Medicago lateral root density. Whereas hydroxylated CEP1 and CEP9 decreased lateral root density, the non-hydroxylated versions did not show any effect compared to the control (Figure 1A and 1B).
Boschiero, Dai and colleagues provide several case studies for the use of the database. For the example of CEP9 (C-Terminally Encoded Peptide 9), they show how the database helps to identify SSP family members and sequence similarity. CEPs are a highly similar SSP family and CEP1 had been previously shown to inhibit M. truncatula lateral root formation (Imin et al., 2013). The authors found that the CEP9 sequence is very similar to that of CEP1 and predicted that the same residues would require hydroxylation to render the peptide functional. They synthesised CEP1 and CEP9 with and without proline hydroxylation and analysed the effect on Medicago lateral root density. Whereas hydroxylated CEP1 and CEP9 decreased lateral root density, the non-hydroxylated versions did not show any effect compared to the control (Figure 1A and 1B).
Another case study shows the identification of a novel SSP in Medicago using the MtSSPdb. By looking closely at RNA-Seq results, the authors found that a locus containing the PSY (Peptide Containing Sulfated Tyrosine) domain in Medicago actually has two transcripts with different expression patterns. They synthesised one of them, PSY7, and showed that its application affects Medicago primary root length and lateral root density .
The MtSSPdb is an exciting new tool for the SSP community. Other databases, such as PlantSSPdb, SPdb and the Arabidopsis Small Secreted Peptide database have been of great use but rely on either few and non-leguminous species, contain few SSPs or are not up to date (Choo et al., 2005; Lease and Walker, 2006; Ghorbani et al., 2015). MtSSPdb is based on a manually curated genome, which helps to detect the small open reading frames, contains a vast number of known and predicted SSPs in Medicago and includes most if not all available gene expression data for the Medicago SSPs. The SSP prediction tool is specialised for the rapidly evolving and short sequences of SSPs and will detect SSPs where other tools might not have been able to. While MtSSPdb is currently limited mostly to Medicago, the authors expect to broaden the database and include further species. Another way to expand the database is to include additional RNA-Seq analyses in the Gene Expression Atlas as they are published. It will be interesting to see how MtSSPdb as a community resource grows with input from the community that it serves.
Literature cited
De Bang TC, Lundquist PK, Dai X, Boschiero C, Zhuang Z, Pant P, Torres-Jerez I, Roy S, Nogales J, Veerappan V, et al (2017) Genome-wide identification of medicago peptides involved in macronutrient responses and nodulation. Plant Physiol 175: 1669–1689
Breiden M, Simon R (2016) Q&A: How does peptide signaling direct plant development? BMC Biol. doi: 10.1186/s12915-016-0280-3
Choo KH, Tan TW, Ranganathan S (2005) SPdb – A signal peptide database. BMC Bioinformatics 6: 249
Czyzewicz N, Yue K, Beeckman T, De Smet I (2013) Message in a bottle: small signalling peptide outputs during growth and development. doi: 10.1093/jxb/ert283
Djordjevic MA, Mohd-Radzman NA, Imin N (2015) Small-peptide signals that control root nodule number, development, and symbiosis. J Exp Bot 66: 5171–5181
Ghorbani S, Lin Y-C, Parizot B, Fernandez A, Njo MF, Van De Peer Y, Beeckman T, Hilson P (2015) Expanding the repertoire of secretory peptides controlling root development with comparative genome analysis and functional assays. J Exp Bot 66: 5257–5269
Imin N, Mohd-Radzman NA, Ogilvie HA, Djordjevic MA (2013) The peptide-encoding CEP1 gene modulates lateral root and nodule numbers in Medicago truncatula. J Exp Bot 64: 5395–409
Lease KA, Walker JC (2006) The Arabidopsis unannotated secreted peptide database, a resource for plant peptidomics. Plant Physiol 142: 831–838
Matsubayashi Y (2014) Posttranslationally Modified Small-Peptide Signals in Plants. Annu Rev Plant Biol 65: 385–413