Modus operandi of Nod-independent symbiosis in Aeschynomene evenia (Plant Physiol)
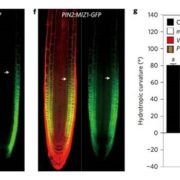
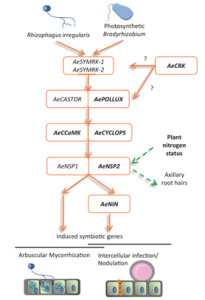 Symbioses and nodule organogenesis processes that occur independently of Nod-factors are relatively unexplored. Quilbe et al. investigated the semi-aquatic legume Aeschynomene evenia, which has recently been shown to establish symbioses with Bradyrhizobium sp. that do not produce Nod factors. The authors used an EMS mutagenesis approach to generate mutations in genes known to be involved in the Nod-factor symbiotic pathway: AePOLLUX, Ca2+/calmodulin dependent kinase (AeCCaMK), AeCYCLOPS, nodulation signaling pathway 2 (AeNSP2), and nodule inception (NIN), as well as a symbiosis-associated cysteine-rich receptor-like kinase (AeCRK). Nodulation kinetics and early nodulin gene expression of the symbiosis mutants were significantly altered in comparison to wild type. For example, pollux-2, pollux-3, pollux-4, cyclops-2 and nin mutants (except nin1) did not produce nodules, while nin-1, pollux-1 and pollux-5 produced underdeveloped nodules with low or no nitrogenase activity. Mycorrhizal colonization and expression of mycorrhization markers were truncated in the roots of the pollux-2, ccamk-3, and cyclops-2 mutants. Axillary root hair crowns, the site of bradyrhizobia colonization, were completely compromised in nsp2 mutants. High nitrate supply suppressed axillary root development and symbiosis associated transcription, while this “nitrogen effect” was lost in nsp2 mutants. These findings show that in Aeschynomene evenia, there is a similar involvement of the Nod signaling pathway throughout the process of rhizobial invasion and symbiosome formation as in other Nod-dependent legumes. (Summary by Lekshmy Sathee @lekshmysnair) Plant Physiol. 10.1093/plphys/kiac325
Symbioses and nodule organogenesis processes that occur independently of Nod-factors are relatively unexplored. Quilbe et al. investigated the semi-aquatic legume Aeschynomene evenia, which has recently been shown to establish symbioses with Bradyrhizobium sp. that do not produce Nod factors. The authors used an EMS mutagenesis approach to generate mutations in genes known to be involved in the Nod-factor symbiotic pathway: AePOLLUX, Ca2+/calmodulin dependent kinase (AeCCaMK), AeCYCLOPS, nodulation signaling pathway 2 (AeNSP2), and nodule inception (NIN), as well as a symbiosis-associated cysteine-rich receptor-like kinase (AeCRK). Nodulation kinetics and early nodulin gene expression of the symbiosis mutants were significantly altered in comparison to wild type. For example, pollux-2, pollux-3, pollux-4, cyclops-2 and nin mutants (except nin1) did not produce nodules, while nin-1, pollux-1 and pollux-5 produced underdeveloped nodules with low or no nitrogenase activity. Mycorrhizal colonization and expression of mycorrhization markers were truncated in the roots of the pollux-2, ccamk-3, and cyclops-2 mutants. Axillary root hair crowns, the site of bradyrhizobia colonization, were completely compromised in nsp2 mutants. High nitrate supply suppressed axillary root development and symbiosis associated transcription, while this “nitrogen effect” was lost in nsp2 mutants. These findings show that in Aeschynomene evenia, there is a similar involvement of the Nod signaling pathway throughout the process of rhizobial invasion and symbiosome formation as in other Nod-dependent legumes. (Summary by Lekshmy Sathee @lekshmysnair) Plant Physiol. 10.1093/plphys/kiac325