Methylome robustness in plants is conferred by a methylation-sensitive system.
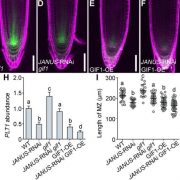
 DNA methylation is instrumental in promoting transcriptional silencing at repetitive elements, inhibiting illegitimate recombination and establishing genomic imprinting. In plants, DNA methylation profiles are stably inherited over generations through the activities of cytosine DNA methyltransferases and 5-methylcytosine glycosylases. Nevertheless, how they interact and influence each other’s activities is not well understood. In this study, Williams and Gehring identify a methylation-sensing gene regulatory system centered on the DNA glycosylase ROS1. ROS1 expression has been described previously as being directly regulated by DNA methylation, and as a consequence it is down-regulated in methylation mutants. By engineering Arabidopsis plants expressing a version of ROS1 insensitive to methylation loss, the authors show that expression of ROS1 in a methylation mutant causes aberrant phenotypes that are amplified over generations. Whole-genome bisulfite sequencing revealed widespread demethylation across the genome. The changes in methylation were not stable through successive generations, with heterochromatin progressively regaining methylation in stark contrast with loci located in euchromatin showing cumulative loss of methylation over generations. Altogether, this study shows that methylation and demethylation pathways are intimately linked within a methylation-sensing system that buffers instability and variations of the methylome. It further supports the idea that the epigenome is particularly robust to destabilizing stimuli. (Summary by Matthias Benoit) Nature Comms. 10.1038/s41467-017-02219-3
DNA methylation is instrumental in promoting transcriptional silencing at repetitive elements, inhibiting illegitimate recombination and establishing genomic imprinting. In plants, DNA methylation profiles are stably inherited over generations through the activities of cytosine DNA methyltransferases and 5-methylcytosine glycosylases. Nevertheless, how they interact and influence each other’s activities is not well understood. In this study, Williams and Gehring identify a methylation-sensing gene regulatory system centered on the DNA glycosylase ROS1. ROS1 expression has been described previously as being directly regulated by DNA methylation, and as a consequence it is down-regulated in methylation mutants. By engineering Arabidopsis plants expressing a version of ROS1 insensitive to methylation loss, the authors show that expression of ROS1 in a methylation mutant causes aberrant phenotypes that are amplified over generations. Whole-genome bisulfite sequencing revealed widespread demethylation across the genome. The changes in methylation were not stable through successive generations, with heterochromatin progressively regaining methylation in stark contrast with loci located in euchromatin showing cumulative loss of methylation over generations. Altogether, this study shows that methylation and demethylation pathways are intimately linked within a methylation-sensing system that buffers instability and variations of the methylome. It further supports the idea that the epigenome is particularly robust to destabilizing stimuli. (Summary by Matthias Benoit) Nature Comms. 10.1038/s41467-017-02219-3