Live-Cell Imaging of Mobile RNAs in Plants
One of the most exciting findings in the past few decades is the discovery that individual mRNAs and noncoding RNAs can act as long-distance signaling messengers traveling cell to cell to distant sites in the plant. Numerous examples unveiled the involvement of endogenous RNAs as non-cell-autonomous regulators of cell fate, organ patterning, and growth responses to environmental signals; other studies even showed that RNAs could be delivered to the next generation (Bayer et al., 2009; Lu et al., 2012; Jiang et al., 2015; Thieme et al., 2015; Martínez et al., 2016). The selectivity and specificity of long-distance transport of endogenous RNAs have been debated intensively, but recent findings indicate that specific cis-acting elements or “zip codes” in the 5′ or 3′ untranslated regions (UTRs) of RNAs are required for RNA mobility (Morris, 2017).
Most of our knowledge of cell-to-cell and long-distance transport in plants originates from studies on how plant viruses manipulate host cells to spread via plasmodesmata and phloem (Ham and Lucas, 2017). Yet, we lack critical information on the transport mechanisms of endogenous RNAs from transcription sites to remote destination cells or tissues. Several RNA biosensors have been successfully used to track the subcellular movement of individual mRNAs in other systems (Mannack et al., 2016); however, these techniques have not been widely used in plants (Tilsner, 2015). One of the most common methods relies on aptamer-protein interactions that exploit the high affinity of transacting RNA-binding proteins to RNA cis-acting elements. The technique consists of engineering two separate genetic constructs. The first contains the MS2 bacteriophage coat protein localized to the nucleus by the canonical Simian Virus 40 (SV40) nuclear localization signal (NLS) fused to GFP (MS2SV40-GFP). The second construct contains an array of MS2 RNA-binding sites, a series of stem-loop (SL) repeats (mRNASL24) inserted in the 5′ or 3′ UTR of the mRNA of interest. Coexpression of these two constructs allows a complex to form between the mRNASL24 and MS2SV40-GFP.
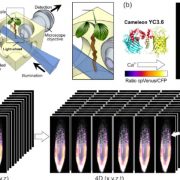
One limitation of this system is the cytosolic background of the MS2-GFP probe; this background masks signals that are due to specific RNA binding. In this issue of Plant Physiology, Luo et al. (2018) successfully adapt the MS2-GFP system to plant cells and examine the subcellular targeting of RNAs in Arabidopsis (Arabidopsis thaliana). Luo et al. replaced NLSSV40, which is inefficiently targeted to the nucleus in plants, with the endogenous nuclear localization signal of the bZIP transcription factor FD (MS2FD-GFP). With the improvement in the signal-to-noise ratio of this biosensor, the authors tested the specificity of the system by coexpressing MS2FD‐GFP with the endogenous mobile FLOWERING LOCUS T (FT) mRNA. They also used a control mRNA that encodes a scorable cytoplasmic marker (RFP). They tagged both constructs with 24 MS2 SL repeats in the 3′ UTR (FTSL24 and RFPSL24). In this setting, both the engineered mRNAs are translocated to the cytoplasm, but only the functional FTSL24-MS2FD-GFP is then actively transported along a Golgi-associated pathway to plasmodesmata (Fig. 1). Although not addressed directly, intrinsic localization elements should be involved in conferring mobility to the FT mRNA, subcellular sorting to plasmodesmata and facilitating cell-to-cell transport. These results agree with the idea that plant viruses use the endogenous plant cellular macromolecule machinery for intracellular and intercellular transport.
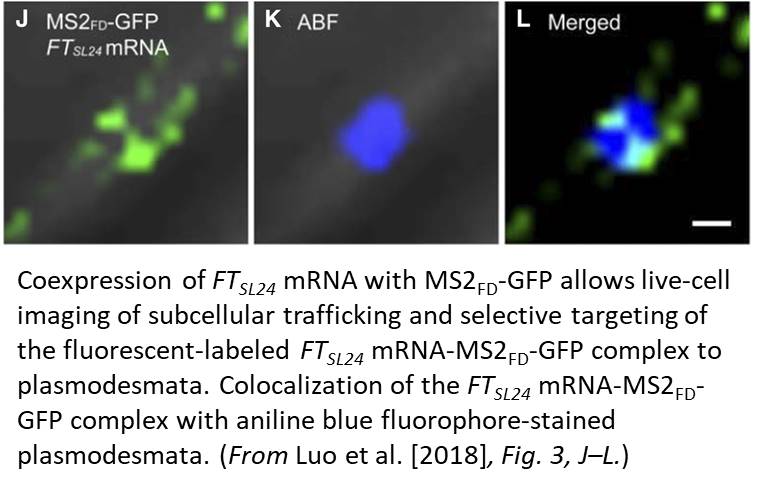
RNA imaging techniques like those presented by Luo et al. (2018) are essential research tools that enable one to examine RNA dynamics in cells in real time. The MS2-GFP system has been the method of choice for quantification of transcription dynamics or examination of subcellular mRNA distribution, which is particularly important in polarized cells, such as pollen tubes or zygotes, to sort polarity or cell fate determinants to specific regions in the cell. Setting the ground for a greater challenge, the fast and extremely stable binding of MS2 to the SL repeats seems well suited for visualization of intercellular and long-distance RNA transport in plants. It remains to be seen if this MS2-GFP system can monitor the subcellular details of RNA trafficking in real time for a wide variety of mobile RNAs. Hopefully, as the research community adopts this useful research tool, those answers will come.
REFERENCES
Bayer M, Nawy T, Giglione C, Galli M, Meinnel T, Lukowitz W (2009) Paternal control of embryonic patterning in Arabidopsis thaliana. Science 323: 1485–1488
Ham BK, Lucas WJ (2017) Phloem-mobile RNAs as systemic signaling agents. Annu Rev Plant Biol 68: 173–195
Jiang H, Yi J, Boavida LC, Chen Y, Becker JD, Köhler C, McCormick S (2015) Intercellular communication in Arabidopsis thaliana pollen discovered via AHG3 transcript movement from the vegetative cell to sperm. Proc Natl Acad Sci USA 112: 13378–13383
Lu KJ, Huang NC, Liu YS, Lu CA, Yu TS (2012) Long-distance movement of Arabidopsis FLOWERING LOCUS T RNA participates in systemic floral regulation. RNA Biol 9: 653–662
Mannack LV, Eising S, Rentmeister A (2016) Current techniques for visualizing RNA in cells. F1000Res 5: 775
Martínez G, Panda K, Köhler C, Slotkin RK (2016) Silencing in sperm cells is directed by RNA movement from the surrounding nurse cell. Nat Plants 2: 16030
Morris RJ (2017) On the selectivity, specificity and signalling potential of the long-distance movement of messenger RNA. Curr Opin Plant Biol 43: 1–7
Thieme CJ, Rojas-Triana M, Stecyk E, Schudoma C, Zhang W, Yang L, Miñambres M, Walther D, Schulze WX, Paz-Ares J, Scheible WR, Kragler F (2015) Endogenous Arabidopsis messenger RNAs transported to distant tissues. Nat Plants 1: 15025
Tilsner J (2015) Techniques for RNA in vivo imaging in plants. J Microsc 258: 1–5