In Brief: Loss of a Silencing Cascade Contributed to Indica Rice Domestication
Thousands of years of artificial selection have produced rice plants (Oryza sativa) that are vastly different from their wild progenitor species in terms of architecture, yield, and resilience. Most of the genetic changes linked to rice domestication involved genes encoding transcription factors. However, gain and loss of microRNAs (miRNAs), the class of small RNAs (21–24 nt) that silence specific target genes, may also have driven rice domestication (Wang et al. 2012).
miRNAs have important functions in plant development, physiology, and stress responses. In some cases, miRNA-mediated silencing is amplified by the production of a fleet of secondary small interfering RNAs (siRNAs) that silence a multitude of mRNAs, a process that requires RNA-DEPENDENT RNA POLYMERASE 6 (RDR6). miRNA length is a strong determinant of this type of silencing cascade; 22-nt miRNAs are sufficient, but not essential, for triggering secondary siRNA production (Manavella et al., 2012).
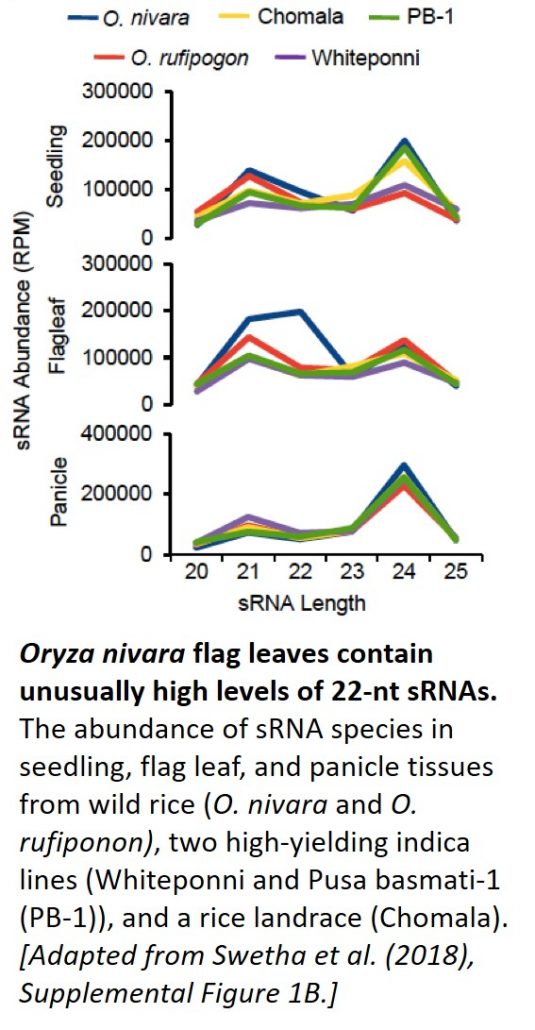 Now, in a Breakthrough Report, Swetha et al. (2018) link miR397 to several of the yield-associated changes that occurred during indica rice domestication. Small RNA profiling of tissues derived from wild rice (O. nivara and O. rufipogon); two high-yielding indica lines; and an assortment of landraces revealed that O. nivara flag leaves had unusually high levels of 22-nt sRNA reads (see figure). The authors mapped these 22-nt reads to MIR397 on chromosome 2. miR397 targets RNAs encoding laccase enzymes, which function in lignification (Lu et al., 2013).
Now, in a Breakthrough Report, Swetha et al. (2018) link miR397 to several of the yield-associated changes that occurred during indica rice domestication. Small RNA profiling of tissues derived from wild rice (O. nivara and O. rufipogon); two high-yielding indica lines; and an assortment of landraces revealed that O. nivara flag leaves had unusually high levels of 22-nt sRNA reads (see figure). The authors mapped these 22-nt reads to MIR397 on chromosome 2. miR397 targets RNAs encoding laccase enzymes, which function in lignification (Lu et al., 2013).
Although miR397 precursors are present in a wide range of plants, their mature form is usually 21-nt long. The additional nucleotide in O. nivara miR397 does not affect targeting ability; psRNA target prediction and tapir analysis under stringent conditions identified 15 laccase targets of 22-nt miR397 in rice. Degradome analysis showed that miR397 targeted at least six laccase transcripts in O. nivara tissues.
The authors next examined whether the 22-nt miR397 abundant in O. nivara flag leaves triggered an RDR6-dependent silencing cascade. The team identified eight regions within O. nivara laccase genes that function as secondary siRNA-producing loci and used a heterologous system to demonstrate that miR397 indeed initiates an RDR6-dependent silencing cascade. As expected, the expression of several laccase genes was substantially reduced in O. nivara and the stems of these wild rice plants accumulated much less lignin than the domesticated rice plants.
Indica lines transgenically expressing MIR397 had reduced lignification and were more similar to O. nivara than to the domesticated lines with respect to yield and panicle, leaf, and stem architecture. Furthermore, twenty-six uncharacterized QTLs previously linked to rice yield overlapped with laccase and MIR397 loci. Thus, the loss of the silencing cascade initiated by miR397 appears to have contributed to key yield-related traits associated with indica rice domestication.
References
Lu, S., Lib, Q., Weid, H., Chang, M.-J., Tunlaya-Anukitb, S., Kimf, H., Liub, J., Songa, Sung, Y.-H., Yuana, L., Yehe, T.-F., Peszlenh. I., Ralphf, J., Sederoffb, R.R., and Chiang, V.L. (2013). Ptr-miR397a is a negative regulator of laccase genes affecting lignin content in Populus trichocarpa. Proc. Natl. Acad. Sci. U. S. A. 110: 10848–10853.
Swetha, C., Basu, D., Pachamuthu, K., Tirumalai, V., Nair, A., Prasad, M., and Shivaprasad, P.V. (2018). Major Domestication-Related Phenotypes in Indica Rice are Due to Loss of miRNA-Mediated Laccase Silencing. Plant Cell; DOI: 10.1105/tpc.18.00472.
Wang, Y., Bai, X., Yan, C., Gui, Y., Wei, X., Zhu, Q.-H., Guo, L., and Fan, L. (2012). Genomic dissection of small RNAs in wild rice (Oryza rufipogon): lessons for rice domestication. New Phytol. 196: 914–925.