Here, there and everywhere: Plastid- and nuclear-localized WHIRLY1 regulates salicylic acid homeostasis during developmental senescence
Does a new job always come with a new location? Perhaps this is true for some plant proteins; half of the proteins are located in more than one subcellular compartment. Emerging evidence in plants shows that nuclear-encoded proteins undergo redox and posttranslational modifications or processing events that could initiate changes in their intracellular localization as well as their function (Foyer et al., 2020).
WHIRLY (WHY) proteins are dually localized, plant-specific proteins that are encoded by three genes in Arabidopsis (Arabidopsis thaliana). All three WHY proteins bind single-stranded DNA, and perhaps RNA, in a sequence non-specific manner (Desveaux et al., 2002; Bach-Pages et al., 2020). WHY1 is partitioned between the nucleus and chloroplast (Krause et al., 2005; Grabowski et al., 2008); however, the mechanisms that might determine its distribution in the cell are only starting to emerge (Ren et al., 2017, Lin et al., 2019). In the nucleus, WHY1 controls the expression of transcription factors, defense genes, and senescence genes by binding their promoters (Krupinska et al., 2013;; Ren et al., 2017). In the chloroplast, WHY1 is required to maintain plastid genome stability, redox balance, and plays a role in retrograde signaling (Cappadocia et al., 2010; Lepage et al., 2013). WHY1 regulates various developmental processes such as senescence. Yet, the factors controlling organellar distribution of WHY1 during senescence are poorly defined and it remains unknown how WHY1 senescence-related functions are handled by the plastid and the nuclear isoforms.
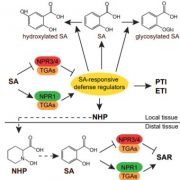
In this issue of Plant Physiology, Lin et al. (2020) further define the function of WHY1 in senescence and explore its role in controlling salicylic acid biosynthesis and conjugation. Analysis of the developmental senescence in why1 mutants showed that WHY1 controls the expression of salicylic acid (SA) biosynthesis genes such as ISOCHORISMATE SYNTHASE (ICS1) and PHENYLALANINE AMMONIA LYASE (PAL) as well as S-ADENOSYL-L-METHIONINE-DEPENDENT METHYLTRANSFERASE (BSMT1), which encodes the protein that generates the mobile signal methyl-SA. Additionally, the authors introduced the why1 mutation into various mutants defective in SA synthesis or signaling, and analyzed the double mutant phenotypes. The outcomes of these experiments allowed them to confirm genetically that WHY1 affects SA levels mainly by controlling PAL1 expression.
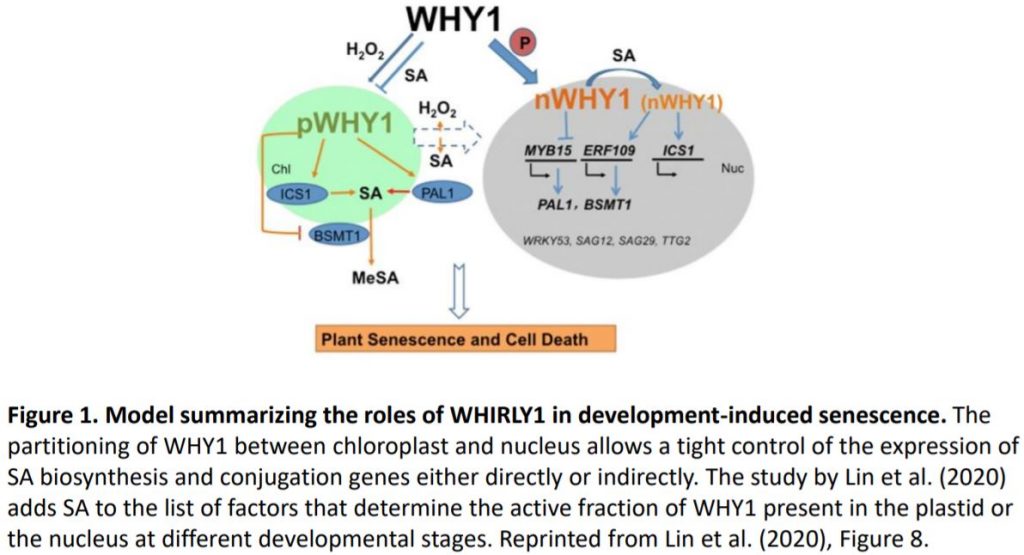 To establish which of the two WHY1 isoforms affects SA metabolism, the authors complemented why1 with plastid, nuclear, or the full-length WHY1 isoforms (Lin et al. 2019). Full-length WHY1 has both a plastid-target peptide for plastid import and a nuclear localization signal. The authors previously engineered a plastid-localized WHY1 by adding a nuclear export sequence to the full-length WHY1, and nuclear WHY1 by eliminating the plastid-target peptide. Complementation with nuclear or full-length WHY1 restored (partially or fully) SA levels and the expression of SA genes to levels comparable of those observed in the wild type. Surprisingly, complementation with plastid WHY1 resulted in the accumulation of SA and increased expression of PAL1 and ICS1, but reduced expression of BSMT1. Altogether these data support the role of nuclear WHY1 in suppressing SA accumulation via suppression of PAL1. However, the authors suggest that plastid WHY1 might induce SA accumulation via repressing BSMT1 during early development and promoting ICS1 at later developmental stages (Figure 1).
To establish which of the two WHY1 isoforms affects SA metabolism, the authors complemented why1 with plastid, nuclear, or the full-length WHY1 isoforms (Lin et al. 2019). Full-length WHY1 has both a plastid-target peptide for plastid import and a nuclear localization signal. The authors previously engineered a plastid-localized WHY1 by adding a nuclear export sequence to the full-length WHY1, and nuclear WHY1 by eliminating the plastid-target peptide. Complementation with nuclear or full-length WHY1 restored (partially or fully) SA levels and the expression of SA genes to levels comparable of those observed in the wild type. Surprisingly, complementation with plastid WHY1 resulted in the accumulation of SA and increased expression of PAL1 and ICS1, but reduced expression of BSMT1. Altogether these data support the role of nuclear WHY1 in suppressing SA accumulation via suppression of PAL1. However, the authors suggest that plastid WHY1 might induce SA accumulation via repressing BSMT1 during early development and promoting ICS1 at later developmental stages (Figure 1).
The above findings are supported by genome-wide analysis of gene expression. Transcriptome mining demonstrated that WHY1 controls numerous hormonal pathways, but the most affected were genes encoding key components of SA metabolism, SA signaling or senescence. Using transactivation assays, Lin et al. also showed that WHY1 activates ICS1 expression directly by binding to its promoter but represses the expression of PAL1, PAL2 and BSMT1 indirectly via MYB15 and ERF105.
Several factors have been shown to affect the organellar distribution of WHY1 including cellular redox state (H2O2 signals) and the WHY1 phosphorylation state (Ren et al., 2017; Lin et al., 2019). In this study, Lin et al. added SA to the list (Figure 1). Methyl-SA treatment was shown to impact WHY1 by reducing the accumulation in the plastids. Furthermore, methyl-SA alters the form of nuclear WHY1 by increasing the accumulation of the small isoform and reducing the accumulation of the large isoform (29 kDa and 37 kDa) respectively. The data also suggest that the status of the nuclear isoforms is dependent on the ICS1 pathway and SA, even though, the nature of the modification induced by SA is still ambiguous.
Overall the data presented by Lin and co-authors demonstrate that plastid and nuclear WHY1 isoforms function concurrently to fine-tune SA metabolism, thus regulating plant senescence (Figure 1). Are the other WHY proteins (WHY2 and WHY3) part of this complex regulation of developmental senescence or do these regulate different processes? Very recently, Huang and coauthors (2020) demonstrated that WHY2 is targeted to chloroplast, mitochondria and the nucleus and is involved in regulating leaf senescence via carbon allocation. In contrast to WHY1, WHY2 functions in the chloroplast remain elusive. Consequently, it seems that dual targeting of proteins to the nucleus and to organelles is a tactic that plants employ to ensure plasticity, organelle communications and retrograde signaling and to be ‘Here and There’.
Amna Mhamdi
Ghent University, Department of Plant Biotechnology and Bioinformatics,
and VIB Center for Plant Systems Biology, 9052 Ghent, Belgium
References
Bach-Pages M, Homma F, Kourelis J, Kaschani F, Mohammed S, Kaiser, M, van der Hoorn RAL, Castello A, Preston GM. (2020) Discovering the RNA-binding proteome of plant leaves with an improved RNA interactome capture method. Biomolecules 10 : 661.
Cappadocia L, Maréchal A, Parent JS, Lepage E, Sygusch J, Brisson N. (2010). Crystal structures of DNA-Whirly complexes and their role in Arabidopsis organelle genome repair. Plant Cell 22:1849-67
Desveaux D, Allard J, Brisson N, Sygusch J (2002) A new family of plant transcription factors displays a novel ssDNA-binding surface. Nat Struct Mol Biol 9, 512-517
Foyer CH, Baker A, Wright M, Sparkes IA, Mhamdi A, Schippers JHM, Van Breusegem F (2020) On the move: redox-dependent protein relocation in plants. JXB. 71: 620-63
Grabowski E, Miao Y, Mulisch M, Krupinska K (2008) Single-stranded DNA-binding protein Whirly1 in barley leaves is located in plastids and the nucleus of the same cell. Plant Physiol 147: 1800-1804
Huang C, Yu J, Cai Q, Chen Y, Li Y, Ren Y, Miao Y (2020) Triple-localized WHIRLY2 influences leaf senescence and silique development via Ccarbon allocation. Plant Physiol. 184:1348-1362.
Krause K, Kilbienski I, Mulisch M, Rodiger A, Schafer A, Krupinska K (2005) DNA-binding proteins of the Whirly family in Arabidopsis thaliana are targeted to the organelles. FEBS letters 579:3707-3712.
Krupinska K, Dähnhardt D, Fischer-Kilbienski I, Kucharewicz W, Scharrenberg C, Trösch M, Buck F (2013) Identification of WHIRLY1 as a factor binding to the promoter of the stress- and senescence-associated Gene HvS40. J. Plant Growth Reg 33: 91-105
Lepage É, Zampini É, Brisson N (2013) Plastid genome instability leads to reactive oxygen species production and plastid-to-nucleus retrograde signaling in Arabidopsis. Plant Physiol. 163 :867-881
Lin W, Huang D, Shi X, Deng B, Ren Y, Lin W Miao Y (2019) H2O2 as a Feedback Signal on Dual-Located WHIRLY1 Associates with Leaf Senescence in Arabidopsis. Cells 8:1585
Lin W, Huang D, Shi X, Deng B, Ren Y, Lin W Miao Y (2020) Dual-localized WHIRLY1 affects salicylic acid biosynthesis via coordination of ISOCHORISMATE SYNTHASE1, PHENYLALANINE AMMONIA LYASE1 and S-ADENOSYL-L-METHIONINE-DEPENDENT METHYLTRANSFERASE1. Plant Physiol. https://doi.org/10.1104/pp.20.00964
Ren Y, Li Y, Jiang Y, Wu B, Miao Y (2017) Phosphorylation of WHIRLY1 by CIPK14 shifts its localization and dual functions in Arabidopsis. Mol Plant 10: 749-763