Genome-wide transcription factor binding in leaves from C3 and C4 grasses (Plant Cell)
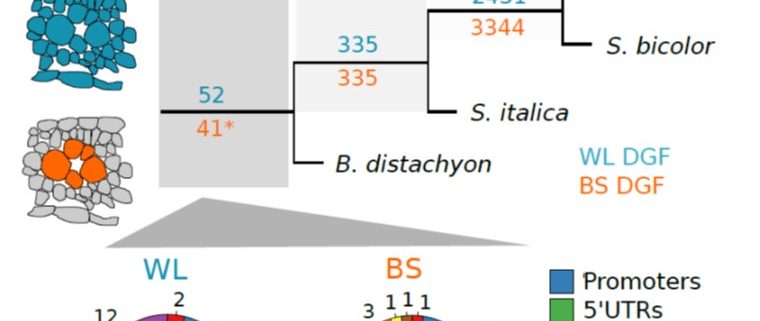
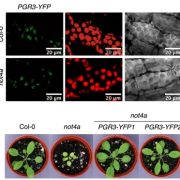
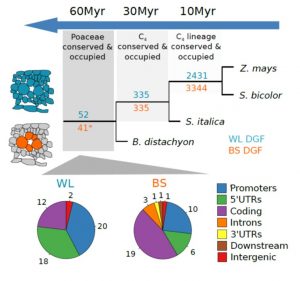 Most plants use the C3 photosynthesis pathway, however many have evolved strategies like C4 photosynthesis that accumulate CO2 around RuBisCO. Burgess et al. performed DNAseI-SEQ in three C4 plants: S. bicolor, Z. mays and S. italica, and one C3: B. dystachion, to offer an insight into the cis-element architecture associated with C3 and C4 photosynthesis. Most of the putative transcription factor (TF) binding sites found were located in gene promoters, however some percentage was located in the exon region. These sites, called duons (dual-use codons), could regulate not only gene expression but also amino acid sequence. The presence of duons was especially high in bundle-sheath cells. For all four grasses, the photosynthesis-gene prevalent cis-elements were AP2-EREBP and MYB classes; however, when orthologous genes were compared most of the TF binding sites were not conserved. Moreover, if cis-elements were conserved their position and frequency varied. (Summary by Cecilia Vasquez-Robinet) Plant Cell 10.1105/tpc.19.00078
Most plants use the C3 photosynthesis pathway, however many have evolved strategies like C4 photosynthesis that accumulate CO2 around RuBisCO. Burgess et al. performed DNAseI-SEQ in three C4 plants: S. bicolor, Z. mays and S. italica, and one C3: B. dystachion, to offer an insight into the cis-element architecture associated with C3 and C4 photosynthesis. Most of the putative transcription factor (TF) binding sites found were located in gene promoters, however some percentage was located in the exon region. These sites, called duons (dual-use codons), could regulate not only gene expression but also amino acid sequence. The presence of duons was especially high in bundle-sheath cells. For all four grasses, the photosynthesis-gene prevalent cis-elements were AP2-EREBP and MYB classes; however, when orthologous genes were compared most of the TF binding sites were not conserved. Moreover, if cis-elements were conserved their position and frequency varied. (Summary by Cecilia Vasquez-Robinet) Plant Cell 10.1105/tpc.19.00078