Plant Science Research Weekly: Sept. 13
Review: Revolutions in agriculture chart a course for targeted breeding of old and new crops ($)
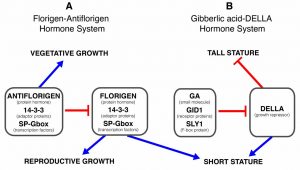 A few traits are associated with domestication across many species. Eshed and Lippman provide an overview of the changes to plant stature and flowering time that have been repeatedly selected by our ancestors. By comparing the molecular underpinnings of these traits across crops, it becomes clear that many different genes can contribute to the desired phenotypes; for example, florigens and antiflorigens have been subjects for selection, as have genes affecting different parts of the gibberellin signaling and respones pathway. In addition to large-effect domestication genes, ongoing breeding has optimized traits particularly for geographical regions through selection on small-effect, modifying genes. The authors point to the potential of gene editing to not only improve crop resiliency to climate change, but also to rapidly improve underutilized crops (such as teff) through alterations in architecture and flowering time. (Summary by Mary Williams) Science 10.1126/science.aax0025 (Note: these topics are also discussed in a recent Teaching Tool in Plant Biology, “Molecular control of plant shoot architecture” written by Agustin Zsögön and Lázaro Eustáquio Pereira Peres).
A few traits are associated with domestication across many species. Eshed and Lippman provide an overview of the changes to plant stature and flowering time that have been repeatedly selected by our ancestors. By comparing the molecular underpinnings of these traits across crops, it becomes clear that many different genes can contribute to the desired phenotypes; for example, florigens and antiflorigens have been subjects for selection, as have genes affecting different parts of the gibberellin signaling and respones pathway. In addition to large-effect domestication genes, ongoing breeding has optimized traits particularly for geographical regions through selection on small-effect, modifying genes. The authors point to the potential of gene editing to not only improve crop resiliency to climate change, but also to rapidly improve underutilized crops (such as teff) through alterations in architecture and flowering time. (Summary by Mary Williams) Science 10.1126/science.aax0025 (Note: these topics are also discussed in a recent Teaching Tool in Plant Biology, “Molecular control of plant shoot architecture” written by Agustin Zsögön and Lázaro Eustáquio Pereira Peres).
Genome-wide transcription factor binding in leaves from C3 and C4 grasses
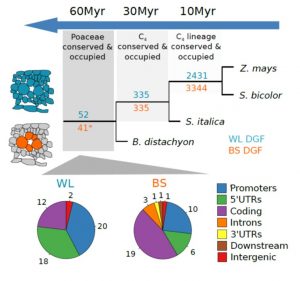 Most plants use the C3 photosynthesis pathway, however many have evolved strategies like C4 photosynthesis that accumulate CO2 around RuBisCO. Burgess et al. performed DNAseI-SEQ in three C4 plants: S. bicolor, Z. mays and S. italica, and one C3: B. dystachion, to offer an insight into the cis-element architecture associated with C3 and C4 photosynthesis. Most of the putative transcription factor (TF) binding sites found were located in gene promoters, however some percentage was located in the exon region. These sites, called duons (dual-use codons), could regulate not only gene expression but also amino acid sequence. The presence of duons was especially high in bundle-sheath cells. For all four grasses, the photosynthesis-gene prevalent cis-elements were AP2-EREBP and MYB classes; however, when orthologous genes were compared most of the TF binding sites were not conserved. Moreover, if cis-elements were conserved their position and frequency varied. (Summary by Cecilia Vasquez-Robinet) Plant Cell 10.1105/tpc.19.00078
Most plants use the C3 photosynthesis pathway, however many have evolved strategies like C4 photosynthesis that accumulate CO2 around RuBisCO. Burgess et al. performed DNAseI-SEQ in three C4 plants: S. bicolor, Z. mays and S. italica, and one C3: B. dystachion, to offer an insight into the cis-element architecture associated with C3 and C4 photosynthesis. Most of the putative transcription factor (TF) binding sites found were located in gene promoters, however some percentage was located in the exon region. These sites, called duons (dual-use codons), could regulate not only gene expression but also amino acid sequence. The presence of duons was especially high in bundle-sheath cells. For all four grasses, the photosynthesis-gene prevalent cis-elements were AP2-EREBP and MYB classes; however, when orthologous genes were compared most of the TF binding sites were not conserved. Moreover, if cis-elements were conserved their position and frequency varied. (Summary by Cecilia Vasquez-Robinet) Plant Cell 10.1105/tpc.19.00078
A reference genome for pea provides insight into legume genome evolution
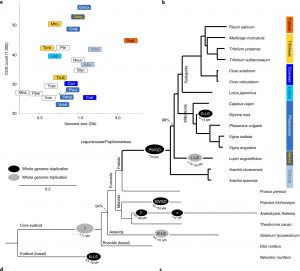 Pea (Pisum sativum) is an important protein-rich crop that introduces most students to the principles of Mendelian genetics, but it has lagged behind in the genomics era due to its much larger genome size than newer model legumes. Kreplak et al. have now released an annotated chromosome-level reference genome assembly for pea. The authors note that the transposon-rich pea genome “has been subject to a higher rate of transposon-mediated unequal recombination giving rise to gain or loss of genes, or ectopic double-strand break repair.” This work provides insights into the evolution of pea and other legumes. The authors reconstruct a paleohistory of modern legumes, explore the genetic relationship of pea varieties, and investigate genes encoding the nutritionally important seed storage proteins, making this a useful resource for breeders. (Summary by Mary Williams) Nature Genetics 10.1038/s41588-019-0480-1
Pea (Pisum sativum) is an important protein-rich crop that introduces most students to the principles of Mendelian genetics, but it has lagged behind in the genomics era due to its much larger genome size than newer model legumes. Kreplak et al. have now released an annotated chromosome-level reference genome assembly for pea. The authors note that the transposon-rich pea genome “has been subject to a higher rate of transposon-mediated unequal recombination giving rise to gain or loss of genes, or ectopic double-strand break repair.” This work provides insights into the evolution of pea and other legumes. The authors reconstruct a paleohistory of modern legumes, explore the genetic relationship of pea varieties, and investigate genes encoding the nutritionally important seed storage proteins, making this a useful resource for breeders. (Summary by Mary Williams) Nature Genetics 10.1038/s41588-019-0480-1
Selaginella moellendorffii expression atlas provides insight into the origin and evolution of plant vasculature
 Land plants (embryophytes) evolved from freshwater algal predecessors over 450 million years ago and have since separated into the morphologically diverse lineages observed today. A key feature in the expansion of plant life on land was the development of the plant vasculature and complex rooting systems. In a new preprint, Ferrari and colleagues present a comprehensive expression atlas for the model early vascular land plant (lycophyte) Selaginella moellendorffii. The authors performed RNA-sequencing analysis to capture genes differentially expressed between the various tissue and cell types of S. moellendorffii. Moreover, a detailed analysis of genes differentially expressed throughout the diurnal (day-night) cycle or upon stress-inducing changes in growth conditions, such as dramatic temperature or light intensity shifts, further revealed the S. moellendorffii transcriptional landscape. To gain an evolutionarily-focused perspective on gene expression across the green plant lineage, the authors performed comparative expression analysis against the CoNekT (Co-expression Network Toolkit) expression database that includes transcriptomes of algae and land plants. This provided new insights into the origin and evolution of secondary metabolism and lignocellulose biosynthesis critical for plant vasculature development, and further delineated the genetic requirements for root development in vascular plants. Collectively, the evolutionary analysis and expression network resources provided by Ferrarri et al. provide an unprecedented look into the form, function, and life history of vascular plants. (Summary by Phil Carella) bioRxiv
Land plants (embryophytes) evolved from freshwater algal predecessors over 450 million years ago and have since separated into the morphologically diverse lineages observed today. A key feature in the expansion of plant life on land was the development of the plant vasculature and complex rooting systems. In a new preprint, Ferrari and colleagues present a comprehensive expression atlas for the model early vascular land plant (lycophyte) Selaginella moellendorffii. The authors performed RNA-sequencing analysis to capture genes differentially expressed between the various tissue and cell types of S. moellendorffii. Moreover, a detailed analysis of genes differentially expressed throughout the diurnal (day-night) cycle or upon stress-inducing changes in growth conditions, such as dramatic temperature or light intensity shifts, further revealed the S. moellendorffii transcriptional landscape. To gain an evolutionarily-focused perspective on gene expression across the green plant lineage, the authors performed comparative expression analysis against the CoNekT (Co-expression Network Toolkit) expression database that includes transcriptomes of algae and land plants. This provided new insights into the origin and evolution of secondary metabolism and lignocellulose biosynthesis critical for plant vasculature development, and further delineated the genetic requirements for root development in vascular plants. Collectively, the evolutionary analysis and expression network resources provided by Ferrarri et al. provide an unprecedented look into the form, function, and life history of vascular plants. (Summary by Phil Carella) bioRxiv
Cannabis glandular trichomes alter morphology and metabolite content during flower maturation ($)
 The therapeutic and recreational applications of Cannabis sativa (Cs) attract public and clinical interest thanks to its secondary metabolites (cannabinoids and terpenes) synthesized and stored in glandular trichomes, hair-like epidermal protrusion densely concentrated in the flowers. Little is known about the properties and contributions of the three different types of glandular trichomes present on the female flower: bulbous, sessile and stalked. The stalked produces more total cannabinoids than sessile, due to the differences in trichome head diameter. Livingston and quilichini et al. described the internal anatomy, biochemistry and transcriptome profile of the glandular trichomes. They define the metabolite storage cavity of stalked and sessile trichome:
The therapeutic and recreational applications of Cannabis sativa (Cs) attract public and clinical interest thanks to its secondary metabolites (cannabinoids and terpenes) synthesized and stored in glandular trichomes, hair-like epidermal protrusion densely concentrated in the flowers. Little is known about the properties and contributions of the three different types of glandular trichomes present on the female flower: bulbous, sessile and stalked. The stalked produces more total cannabinoids than sessile, due to the differences in trichome head diameter. Livingston and quilichini et al. described the internal anatomy, biochemistry and transcriptome profile of the glandular trichomes. They define the metabolite storage cavity of stalked and sessile trichome:
- Stalked: blue autofluorescence correlate with high cannabinoid levels; 12-16 secretory cells and strongly monoterpene-dominant terpene profiles.
- Sessile: red-shifted autofluorescence; 8 secretory cells and less monoterpene-dominant terpene profile.
Intrinsic autofluorescence patterns and disc cell numbers promoted a developmental model where sessile trichomes may represent a premature stage of stalked trichome formation. Moreover, the metabolic specialization of stalked trichomes for monoterpene production is highlighted by the identification and characterization of two unknown and highly expressed monoterpene synthases: CsTPS37FN and CsTPS38N. (Summarized by Francesca Resentini) Plant Journal 10.1111/tpj.14516
Halotropism requires phospholipase Dζ1‐mediated modulation of cellular polarity of auxin transport carriers ($)
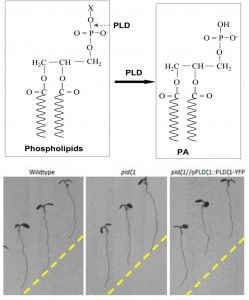 Phospholipase D (PLD) enzymes cleave phosopholipds to release phosphatidic acid (PA), which is a signal that affects membrane dynamics. Previous studies have indicated a role for PLD and PA in root responses to osmotic and salt stress, but as the Arabidopsis genome has 12 genes encoding PLD questions remain about their specific functions. Korver et al. have investigated the role of PLDζ1 (zeta1), which has sequence features resembling mammalian PLDs. They found that knockout mutants show delayed halotropism (bending away from salt) and an exaggerated gravitropic response, as well as abnormalities in the redistribution of the auxin transporter PIN2. Osmotic stress induced membrane structures (OSIMS) that occur shortly after osmotic or salt stress have a prolonged presence in the pldζ1 mutant. The authors conclude that the delayed halotropism response of the pldζ1 mutant is a consequence of reduced auxin asymmetry arising from altered PIN2 distribution. (Summary by Mary Williams) Plant Cell Environ. 10.1111/pce.13646
Phospholipase D (PLD) enzymes cleave phosopholipds to release phosphatidic acid (PA), which is a signal that affects membrane dynamics. Previous studies have indicated a role for PLD and PA in root responses to osmotic and salt stress, but as the Arabidopsis genome has 12 genes encoding PLD questions remain about their specific functions. Korver et al. have investigated the role of PLDζ1 (zeta1), which has sequence features resembling mammalian PLDs. They found that knockout mutants show delayed halotropism (bending away from salt) and an exaggerated gravitropic response, as well as abnormalities in the redistribution of the auxin transporter PIN2. Osmotic stress induced membrane structures (OSIMS) that occur shortly after osmotic or salt stress have a prolonged presence in the pldζ1 mutant. The authors conclude that the delayed halotropism response of the pldζ1 mutant is a consequence of reduced auxin asymmetry arising from altered PIN2 distribution. (Summary by Mary Williams) Plant Cell Environ. 10.1111/pce.13646
Nuclear-cytoplasmic partitioning of ARF proteins controls auxin responses in Arabidopsis thaliana
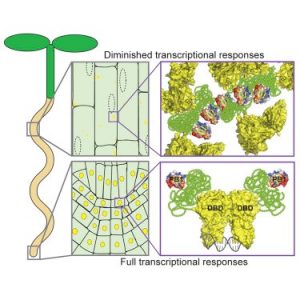 ARF (Auxin Response Factor) transcription factor proteins contain three domains: an N-terminal DNA-binding domain, a variable middle region, and a C-terminal PB1 domain. Powers et al. found that ARF7 and ARF19 accumulate as large-order assemblies in the cytoplasmic region of mature root cells, but the cytoplasmic signal disappears in the actively dividing cells. They showed that a mutation in the PB1 domain leads to nuclear instead of cytoplasmic localization and interferes with assembly formation. The authors also identified prion-like domains within the middle region that contribute to protein condensate formation. Disruption of ARF condensate formation results in altered transcription and morphological defects. This study provides a model where ARF nucleo-cytoplasmic partitioning and condensate formation regulate auxin response and maintains morphological features. (Summary by Arif Ashraf) Molecular Cell 10.1016/j.molcel.2019.06.044
ARF (Auxin Response Factor) transcription factor proteins contain three domains: an N-terminal DNA-binding domain, a variable middle region, and a C-terminal PB1 domain. Powers et al. found that ARF7 and ARF19 accumulate as large-order assemblies in the cytoplasmic region of mature root cells, but the cytoplasmic signal disappears in the actively dividing cells. They showed that a mutation in the PB1 domain leads to nuclear instead of cytoplasmic localization and interferes with assembly formation. The authors also identified prion-like domains within the middle region that contribute to protein condensate formation. Disruption of ARF condensate formation results in altered transcription and morphological defects. This study provides a model where ARF nucleo-cytoplasmic partitioning and condensate formation regulate auxin response and maintains morphological features. (Summary by Arif Ashraf) Molecular Cell 10.1016/j.molcel.2019.06.044
Evolution of the KANADI gene family and leaf development in lycophytes and ferns ($)
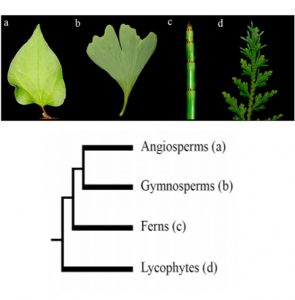 Plants constantly grow post embryonically through shoot and root apical meristems. Multiple transcriptional factors and other regulators fine-tune such growth. Leaf growth from the apical meristem is well studied in the model plant Arabidopsis where two main transcription factors, namely HD-ZIP III (Class III Homeodomain Leucine Zipper protein) and KANADI (GARP Family proteins), play key roles. The adaxial expression of HD-ZIP III and abaxial expression of KANADI genes help maintain leaf bilateral symmetry. Cardona et al., using phylogenetic and gene expression analysis, determined if the KANADI genes have conserved expression domains like Arabidopsis in lycophytes and ferns. Using the OneKP database and other genome databases, the authors identified KANADI orthologs in specific ferns and lycophytes. Subsequently they determined the expression of KANADI genes in the lycophyte Selaginella moellendotffii and in the fern Equisetum hyemale. They conclude that the KANADI gene expression pattern in the abaxial side is conserved in the fern but not in lycophytes. (Summary by Suresh Damodaran) Plants 10.3390/plants8090313
Plants constantly grow post embryonically through shoot and root apical meristems. Multiple transcriptional factors and other regulators fine-tune such growth. Leaf growth from the apical meristem is well studied in the model plant Arabidopsis where two main transcription factors, namely HD-ZIP III (Class III Homeodomain Leucine Zipper protein) and KANADI (GARP Family proteins), play key roles. The adaxial expression of HD-ZIP III and abaxial expression of KANADI genes help maintain leaf bilateral symmetry. Cardona et al., using phylogenetic and gene expression analysis, determined if the KANADI genes have conserved expression domains like Arabidopsis in lycophytes and ferns. Using the OneKP database and other genome databases, the authors identified KANADI orthologs in specific ferns and lycophytes. Subsequently they determined the expression of KANADI genes in the lycophyte Selaginella moellendotffii and in the fern Equisetum hyemale. They conclude that the KANADI gene expression pattern in the abaxial side is conserved in the fern but not in lycophytes. (Summary by Suresh Damodaran) Plants 10.3390/plants8090313
teosinte branched1 and the control of bud dormancy and growth in maize
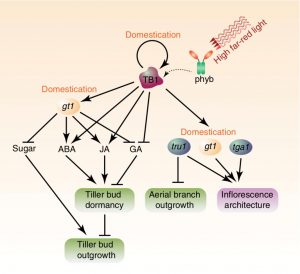 Branching has a significant impact on plant architecture. Maize, a domesticated species, has lower branching levels in comparison to its wild ancestor teosinte. Axillary branching is the result of lateral buds growth. Some factors such as phytohormones and transport of sugars are related to bud activation/dormancy and hence branching. In a recent paper, Dong et al. studied the dynamics of bud dormancy at the transcriptomic level in maize. They compared data from B73 to teosinte branched1 (tb1) and grassy tillers1 (gt1) two mutants that over-produce tillers and aerial branches. As a result, they obtained a set of genes involved in the regulation of bud dormancy. Their further analysis focussed on TB1 and its targets. Among these were genes involved in ABA synthesis and transport, and ABA signal transduction. Other hormonal processes identified included gibberellins; TB1 inhibits GA production and signaling, resulting in bud dormancy. Additionally, some TB1 targets control the sugar distribution; the levels of sucrose, fructose, and related metabolites were elevated in tb1 buds. TB1 also controls other domestication genes, such as tga1 and tru1, that regulates inflorescence architecture. As the authors conclude, “This places tb1 on top of the domestication hierarchy, demonstrating its critical importance during the domestication of maize from teosinte.” (Summary by Humberto Herrera-Ubaldo) Nature Comms. 10.1038/s41467-019-11774-w
Branching has a significant impact on plant architecture. Maize, a domesticated species, has lower branching levels in comparison to its wild ancestor teosinte. Axillary branching is the result of lateral buds growth. Some factors such as phytohormones and transport of sugars are related to bud activation/dormancy and hence branching. In a recent paper, Dong et al. studied the dynamics of bud dormancy at the transcriptomic level in maize. They compared data from B73 to teosinte branched1 (tb1) and grassy tillers1 (gt1) two mutants that over-produce tillers and aerial branches. As a result, they obtained a set of genes involved in the regulation of bud dormancy. Their further analysis focussed on TB1 and its targets. Among these were genes involved in ABA synthesis and transport, and ABA signal transduction. Other hormonal processes identified included gibberellins; TB1 inhibits GA production and signaling, resulting in bud dormancy. Additionally, some TB1 targets control the sugar distribution; the levels of sucrose, fructose, and related metabolites were elevated in tb1 buds. TB1 also controls other domestication genes, such as tga1 and tru1, that regulates inflorescence architecture. As the authors conclude, “This places tb1 on top of the domestication hierarchy, demonstrating its critical importance during the domestication of maize from teosinte.” (Summary by Humberto Herrera-Ubaldo) Nature Comms. 10.1038/s41467-019-11774-w
The QKY-SYP121 complex controls long-distance florigen movement ($)
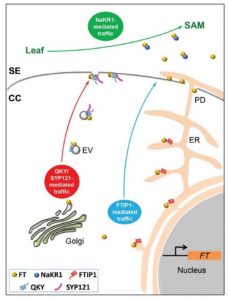 In Arabidopsis thaliana, changes in day-length (photoperiod) activate the expression and transport of phloem-mobile florigen (FT, FLOWERING LOCUS T) to the shoot apical meristem to trigger the transition to flowering. While the role of FT as a long-distance signal is well-established, the underlying mechanisms controlling its movement from phloem companion cells (CCs) to sieve elements (SEs) for access to the phloem translocation stream is not well understood. Liu et al. investigated the role of membrane-resident SNARE proteins controlling endosomal vesicular trafficking for potential roles in regulating the CC-to-SE movement of FT, starting with the observation of a late-flowering phenotype in syp121-4 (syntaxin of plants 121) mutants. They subsequently showed that SYP121 interacts with the MCTP (Multiple C2 domain-containing Transmembrane Protein) family protein QUIRKY (QKY), much like the related MCTP family member, FT-INTERACTING PROTEIN1 (FTIP1). They observed FT movement defects in syp121, qky, and ftip1 mutants relative to wild-type controls. Collectively, this work reveals that a QKY-SYP121 complex mediates FT movement from CCs-to-SEs through endosomal vesicular trafficking at the plasma membrane, which occurs alongside FTIP1-mediated transport. (Summary by Phil Carella) Plant Cell 10.1105/tpc.18.00960
In Arabidopsis thaliana, changes in day-length (photoperiod) activate the expression and transport of phloem-mobile florigen (FT, FLOWERING LOCUS T) to the shoot apical meristem to trigger the transition to flowering. While the role of FT as a long-distance signal is well-established, the underlying mechanisms controlling its movement from phloem companion cells (CCs) to sieve elements (SEs) for access to the phloem translocation stream is not well understood. Liu et al. investigated the role of membrane-resident SNARE proteins controlling endosomal vesicular trafficking for potential roles in regulating the CC-to-SE movement of FT, starting with the observation of a late-flowering phenotype in syp121-4 (syntaxin of plants 121) mutants. They subsequently showed that SYP121 interacts with the MCTP (Multiple C2 domain-containing Transmembrane Protein) family protein QUIRKY (QKY), much like the related MCTP family member, FT-INTERACTING PROTEIN1 (FTIP1). They observed FT movement defects in syp121, qky, and ftip1 mutants relative to wild-type controls. Collectively, this work reveals that a QKY-SYP121 complex mediates FT movement from CCs-to-SEs through endosomal vesicular trafficking at the plasma membrane, which occurs alongside FTIP1-mediated transport. (Summary by Phil Carella) Plant Cell 10.1105/tpc.18.00960
A single amino-acid substitution impairs PTI and ETI in an SA-dependent manner in rice ($)
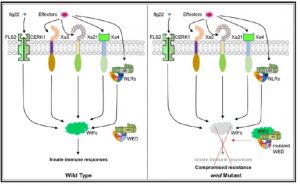 Plants possess two immune strategies to prevent invasion by pathogens called pattern-triggered immunity (PTI, typically mediated by cell-surface receptors) and effector-triggered immunity (ETI). During ETI, intracellular resistance (R) proteins perceive specific pathogen effectors. Tang et al. describe a gain-of-function rice R gene mutant designated as weaker defense (wed), in which disease resistance conferred by both PTI and ETI against Xanthomonas oryzae pv. oryzae (Xoo) is suppressed. These results implied in rice there is an overlap in defense signaling conferred by multiple R genes, and a shared pathway between PTI and ETI. Interestingly, the wed mutants have elevated salicylic acid levels, although defense responses and expression of NPR1 are decreased, suggesting that its effects are SA-independent. In summary, this gain-of-function mutant could be a powerful tool for genetic dissection of shared immune signaling pathways activated by different types of immune receptors. (Summary by Nanxun Qin) Plant Physiology 10.1104/pp.19.00686
Plants possess two immune strategies to prevent invasion by pathogens called pattern-triggered immunity (PTI, typically mediated by cell-surface receptors) and effector-triggered immunity (ETI). During ETI, intracellular resistance (R) proteins perceive specific pathogen effectors. Tang et al. describe a gain-of-function rice R gene mutant designated as weaker defense (wed), in which disease resistance conferred by both PTI and ETI against Xanthomonas oryzae pv. oryzae (Xoo) is suppressed. These results implied in rice there is an overlap in defense signaling conferred by multiple R genes, and a shared pathway between PTI and ETI. Interestingly, the wed mutants have elevated salicylic acid levels, although defense responses and expression of NPR1 are decreased, suggesting that its effects are SA-independent. In summary, this gain-of-function mutant could be a powerful tool for genetic dissection of shared immune signaling pathways activated by different types of immune receptors. (Summary by Nanxun Qin) Plant Physiology 10.1104/pp.19.00686



