Genome assembly and population genomic analysis provide insights into the evolution of modern sweet corn (Nature Comms)
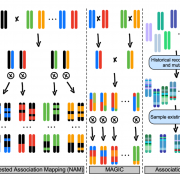
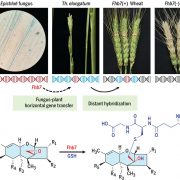
 In maize (Zea mays), loss of function mutations in genes involved in starch biosynthesis characterize sweet corn varieties that have increased sugar content in the kernel. Specifically, lines carrying the shrunken2 (sh2) allele revolutionized the corn industry in Northern America in the last 50 years thanks to improved quality traits and shelf life. Besides the kernel, these cultivars also display several phenotypic alterations compared to field corn, suggesting they originated through divergent selection. In this paper, Hu and coworkers presented a high-quality reference genome of an agronomically important sweet corn inbred line (la435) harboring the sh2-R allele (Ia453-sh2). Results of Transposable Elements (TE) annotation and pairwise genomic comparison with six field corn varieties, including the B73 reference genome, indicated high level of divergence in TE families among accessions, which could be related to genetic variation in protein-coding genes. As an example, the sweet corn la453-sh2 defective allele is associated with structural rearrangements causing a 50 Kb separation between the first and the second half of the gene. This big TE insertion – which is absent in B73 – caused the loss of a conserved domain in the enzyme that controls the first irreversible reaction of starch biosynthesis, leading to higher sugar accumulation in the endosperm. This paper presents novel insights into the phylogenetic relationships among more than 5000 maize accessions and identifies candidate genes likely to control additional sweet corn traits such as leaf architecture (Drooping leaf), short stature (Dwarf1) and early flowering (FT-interacting protein). (Summary and image adaptation by Michela Osnato @michela_osnato). Nature Comms. 10.1038/s41467-021-21380-4
In maize (Zea mays), loss of function mutations in genes involved in starch biosynthesis characterize sweet corn varieties that have increased sugar content in the kernel. Specifically, lines carrying the shrunken2 (sh2) allele revolutionized the corn industry in Northern America in the last 50 years thanks to improved quality traits and shelf life. Besides the kernel, these cultivars also display several phenotypic alterations compared to field corn, suggesting they originated through divergent selection. In this paper, Hu and coworkers presented a high-quality reference genome of an agronomically important sweet corn inbred line (la435) harboring the sh2-R allele (Ia453-sh2). Results of Transposable Elements (TE) annotation and pairwise genomic comparison with six field corn varieties, including the B73 reference genome, indicated high level of divergence in TE families among accessions, which could be related to genetic variation in protein-coding genes. As an example, the sweet corn la453-sh2 defective allele is associated with structural rearrangements causing a 50 Kb separation between the first and the second half of the gene. This big TE insertion – which is absent in B73 – caused the loss of a conserved domain in the enzyme that controls the first irreversible reaction of starch biosynthesis, leading to higher sugar accumulation in the endosperm. This paper presents novel insights into the phylogenetic relationships among more than 5000 maize accessions and identifies candidate genes likely to control additional sweet corn traits such as leaf architecture (Drooping leaf), short stature (Dwarf1) and early flowering (FT-interacting protein). (Summary and image adaptation by Michela Osnato @michela_osnato). Nature Comms. 10.1038/s41467-021-21380-4