Field of Genes: Uncovering EGRINs (Environmental Gene Regulatory Influence Networks) in Rice That Function during High-Temperature and Drought Stress
IN BRIEF by Jennifer Lockhart [email protected]
Heat and drought stress greatly restrict crop productivity, but most of what we know about a plant’s response to these stresses comes from controlled laboratory studies. This factor, along with the complex nature of these responses, has hampered efforts to breed and engineer crops with improved stress tolerance. Plants respond to fluctuating environments through the altered expression of numerous genes whose transcription is tightly coordinated. Enter the EGRINs (environmental gene regulatory influence networks), which orchestrate the timing and rate of transcription of these genes in response to environmental signals. Transcription factors (TFs), comprising the core of EGRINs, interact with conserved regulatory elements in genomic DNA to modulate their expression. Since plants can’t run away from their environments, they likely have scores of EGRIN modules that help them cope with changing environments. To date, most EGRINs have been identified using algorithms based on transcriptome data under the assumption that gene expression patterns can be used to predict the relationships between TFs and the genes they regulate. However, simply identifying groups of genes that are coexpressed under stress conditions fails to capture the complexity of these interactions. Since EGRINs affect chromatin availability in multiple regions of the genes they regulate, Sullivan et al. (2014) used changes in chromatin accessibility and the identification of transcription factor footprints, rather than changes in gene expression, to identify putative stress-responsive transcriptional networks in Arabidopsis thaliana.
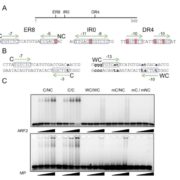
Wilkins et al. (2016) went one step further by combining chromatin accessibility analysis with coexpression data and network inference algorithms to uncover EGRINs in drought- and heat-stressed rice (Oryza sativa), a major crop whose yields are threatened by climate change. Specifically, the authors investigated the responses of five tropical Asian rice cultivars to high-temperature and water-stress conditions using both hydroponically grown plants in a controlled environment and plants that were naturally exposed to stress conditions in the field (see figure). The authors performed time-series transcriptome analysis (RNA-seq) and chromatin accessibility analysis (ATAC-seq; assay of transposase accessible chromatin) to connect TFs with genes containing accessible, known TF binding motifs in their promoters in order to organize what is currently known about putative regulatory interactions. This analysis uncovered 5447 putative target genes for 445 transcription factors. After various filtering steps, the authors performed network component analysis, a technique used to build upon known networks based on output such as gene expression data (Liao et al., 2003). Final networks were deduced using a network-learning tool known as the Inferelator algorithm (Greenfield et al., 2013).

This analysis uncovered possible EGRINs involving 4052 target genes regulated by 113 TFs. Most TFs appear to regulate target genes in response to a single stress treatment, whereas a small number of TFs regulate their targets under multiple stress conditions. The use of data from both growth chamber- and field-grown plants allowed the authors to infer EGRINs that function in response to both rapid, controlled stress conditions and fluctuating environments in the field, producing an unusually rich set of data. The EGRINs inferred in this study could be refined and may be used someday to identify high-priority targets for plant breeding and biotechnology programs, fulfilling the dreams of biologists and farmer alike.
REFERENCES
Greenfield, A., Hafemeister, C., and Bonneau, R. (2013). Robust data-driven incorporation of prior knowledge into the inference of dynamic regulatory networks. Bioinformatics 29: 1060–1067.
Liao, J.C., Boscolo, R., Yang, Y.-L., Tran, L.M., Sabatti, C., and Roychowdhury, V.P. (2003). Network component analysis: Reconstruction of regulatory signals in biological systems. Proc. Natl. Acad. Sci. USA 100: 15522–15527.
Sullivan, A.M., et al. (2014). Mapping and dynamics of regulatory DNA and transcription factor networks in A. thaliana. Cell Reports 8: 2015–2030.
Wilkins, O., Hafemeister, C., Plessis, A., Holloway-Phillips, M.-M., Pham, G.M., Nicotra, A.B., Gregorio, G.B., Jagadish, S.V.K., Sptiningsih, E.M., Bonneau, R., and Purugganan, M. (2016). EGRINs (environmental gene regulatory influence networks) in rice that function in the response to water deficit, high temperature, and agricultural environments. Plant Cell 28: 2365–2384.






Leave a Reply
Want to join the discussion?Feel free to contribute!