Dynamic Alternative Splicing Response to Cold
Calixto et al. demonstrate a rapid wave of alternative splicing as temperatures drop. The Plant Cell (2018) https://doi.org/10.1105/tpc.18.00177.
By Cristiane Calixto and John Brown
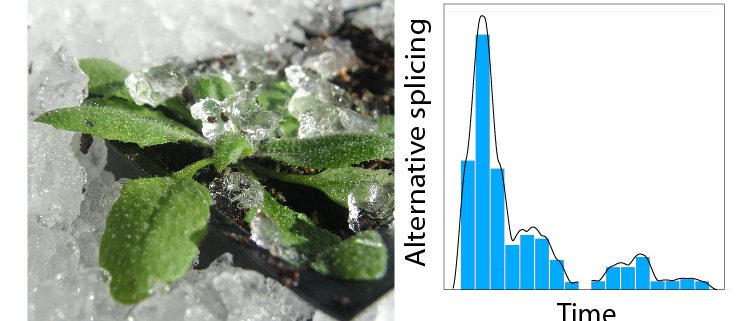
Background: Alternative splicing (AS) allows a single gene to produce more than one transcript, which can affect protein production and function. AS is common and essential for eukaryotes to control gene expression. The scale and dynamics of AS in plant stress responses has never been addressed but is important because of expected increases in environmental stresses due to climate change. We have exploited new tools and methods that we developed, including a comprehensive transcript database for Arabidopsis and analysis of time-series RNA-sequencing (RNA-Seq) data, to examine accurately changes in expression and AS at the gene and transcript levels.
Question: To what extent does AS affect gene expression in plants during exposure to low temperature?
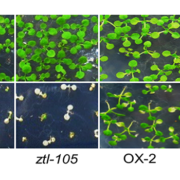
Findings: We found a massive and rapid wave of AS coincident with the transcriptional response. Dynamic analysis identified hundreds of genes with drastically altered AS in the first few hours of cold—“early AS” genes—and included many transcription and splicing factor regulators that had not been identified previously as cold responsive. We demonstrated the speed and sensitivity of AS of some of these genes to small temperature reductions such that plants may fine-tune expression dynamically via AS. Some of the early AS splicing factor genes are needed for plants to acclimate to low temperatures and survive freezing. They likely drive cascades of AS of downstream genes parallel to and coordinated with the transcriptional response. We also provide a step-change in analysis of RNA-seq data and AS.
Next steps: We are studying the functions of splicing and transcription factors that are regulated by AS and low temperature and identifying their target genes and mechanisms of regulation. The AS contribution to the cold response is likely to also occur in responses to different stresses and to pests and pathogens. The new tools and methods we developed will advance our understanding of gene expression regulation under stress, which in view of climate change will play a key role in understanding crop responses to the environment.
Cristiane P. G. Calixto, Wenbin Guo, Allan B. James, Nikoleta A. Tzioutziou, Juan Carlos Entizne, Paige E. Panter, Heather Knight, Hugh G. Nimmo, Runxuan Zhang and John W. S. Brown. (2018) Rapid and dynamic alternative splicing impacts the Arabidopsis cold response transcriptome. Plant Cell DOI: https://doi.org/10.1105/tpc.18.00177.