Divide and Conquer: Introducing a Novel Player in Cell Plate Formation
IN BRIEF by Kathleen L. Farquharson [email protected]
Polysaccharide-rich cell walls are a distinguishing feature of plants that influence many aspects of growth and development, including cell division. Whereas contractile rings pinch dividing cells into two daughter cells in other eukaryotes, newly built cell walls partition the products of plant cell division. The phragmoplast guides construction of a cell plate at the center of the division plane, which expands outward from the cell center until it fuses with the parental cell wall, completely separating the daughter cells. As cellulose is the major load-bearing component of cell walls, cellulose synthase (CESA) and cellulose synthase-like (CSL) enzymes, which have a high degree of sequence identity with CESA and are thought to catalyze the biosynthesis of cell wall polysaccharides, are expected to be involved in the construction of new cell walls.
To identify genes that are specifically required for de novo cell wall synthesis in dividing cells, Gu et al. (2016) mined transcriptional profiles of an Arabidopsis thaliana stomatal lineage. They found that three CSLD genes, CSLD2, 3, and 5, were highly expressed in early stage stomatal cells. In contrast to CSLD2 and 3, CSLD5 was strongly expressed in rapidly dividing cell populations and weakly expressed in expanding and differentiating cells. Consistent with a role in cell division, chromatin immunoprecipitation indicated that CSLD5 is a direct target of SPEECHLESS (SPCH), a master transcriptional regulator of divisions during stomatal development (Lau et al., 2014). Mutant csld5 plants exhibited defects in growth and development. In addition to having reduced stature and shorter roots and smaller rosettes than wild-type plants (as reported in Bernal et al., 2007; Yin et al., 2011), the csld5 mutant displayed aberrant stomatal clusters and cell wall stubs in both aerial and root tissues. Although csld2 and 3 did not exhibit these defects, loss of either CSLD2 or 3 enhanced the csld5 phenotype, indicating functional redundancy among CSLD genes.
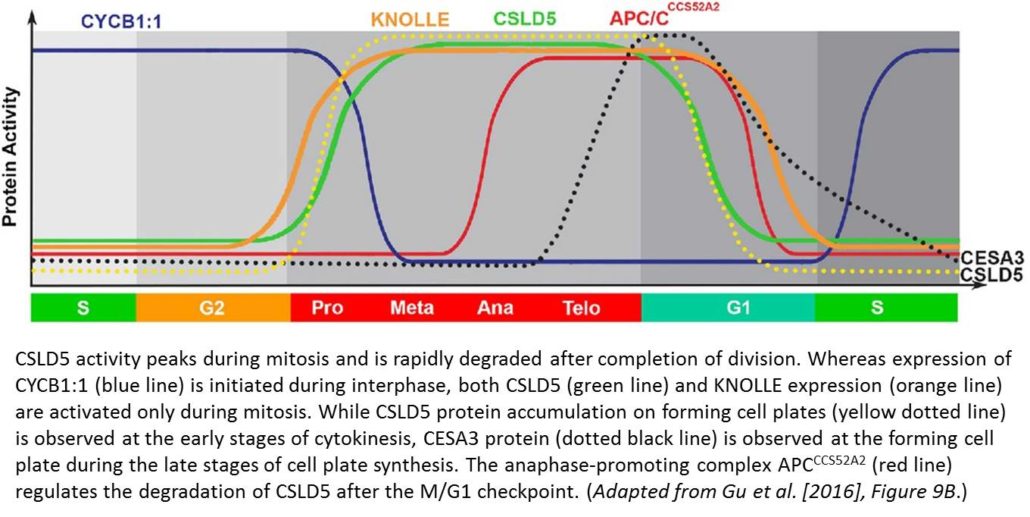
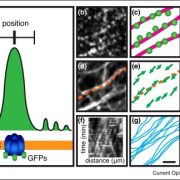
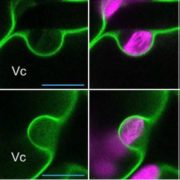
Fluorescently tagged CSLD2, 3, and 5 all localized to the cell plates of dividing cells. However, whereas CSLD2 and CSLD3 also accumulated in punctate structures of nondividing cells, CSLD5 was preferentially associated with actively dividing cells. A comparison of the distribution of fluorescently tagged CSLD5 with that of fluorescently tagged CYCB1:1 (a cyclin) and CESA3 and KNOLLE (two other cell plate-localized proteins) showed that CSLD5 started to accumulate during early mitosis and vanished soon after completion of cytokinesis (see figure). The authors demonstrated that the rapid loss of CSLD5 was due to ubiquitination by the anaphase-promoting complex (APC).
This work establishes CSLD5 as a cell cycle-regulated enzyme that deposits cell wall material in expanding cell plates during cytokinesis. The next challenge is to identify which compound CSLD5 adds to the cell plate and to pinpoint the individual contributions of CSLD proteins and other cell plate-localized enzymes to the construction of sturdy walls during plant cell division.
 REFERENCES
REFERENCES
Bernal, A.J., Jensen, J.K., Harholt, J., Sørensen, S., Moller, I., Blaukopf, C., Johansen, B., de Lotto, R., Pauly, M., Scheller, H.V., Willats, W.G.T. (2007). Disruption of ATCSLD5 results in reduced growth, reduced xylan and homogalacturonan synthase activity and altered xylan occurrence in Arabidopsis. Plant J. 52: 791–802.
Gu, F., Bringmann, M., Combs, J., Yang, J., Bergmann, D., Nielsen, E. (2016). Arabidopsis CSLD5 functions in cell plate formation in a cell cycle-dependent manner. Plant Cell 28: 1722–1737.
Lau, O.S., Davies, K.A., Chang, J., Adrian, J., Rowe, M.H., Ballenger, C.E., Bergmann, D.C. (2014). Direct roles of SPEECHLESS in the specification of stomatal self-renewing cells. Science 345: 1605–1609.
Yin, L., Verhertbruggen, Y., Oikawa, A., Manisseri, C., Knierim, B., Prak, L., Jensen, J.K., Knox, J.P., Auer, M., Willats, W.G.T., Scheller, H.V. (2011). The cooperative activities of CSLD2, CSLD3, and CSLD5 are required for normal Arabidopsis development. Mol. Plant 4: 1024–1037.



Leave a Reply
Want to join the discussion?Feel free to contribute!