Divide and Conquer: High-Throughput Screening of Chlamydomonas Cell Cycle Mutants
Cell division is essential for growth and reproduction. The cell cycle machinery is well conserved between yeast and animals, but whether this conservation extends to the plant lineage is not clear, having diverged over two billion years ago: ample time and opportunity for divergence in sequence and function, and for introduction of plant-specific innovations.
Discovering genes involved in the plant cell cycle is no small task, as apparent sequence conservation does not always imply conservation of function (Nowach et al., 2012). Forward genetic screens are less influenced by prior knowledge, but how to devise a mutant screen for essential genes? Breker, Lieberman and Cross (Breker et al., 2018) describe here an updated and optimized pipeline for finding cell cycle genes in the unicellular alga Chlamydomonas reinhardtii.
Chlamydomonas is a member of the plant kingdom, yet offers many of the advantages of a microbe: a haploid (and fully sequenced) genome and fast generation time. Insertional mutagenesis is available, but tends to generate null (and dead) alleles. Constitutive death is not a very useful phenotype. The generation of temperature-sensitive (Ts) alleles is more suitable, but comes at a cost: Ts alleles are rare, so 1) the mutant pool needs to be larger, and 2) all mutants need to be tested under both permissive and restrictive temperatures. Breker and colleagues leaned on robotics to replicate the many, many plates needed.
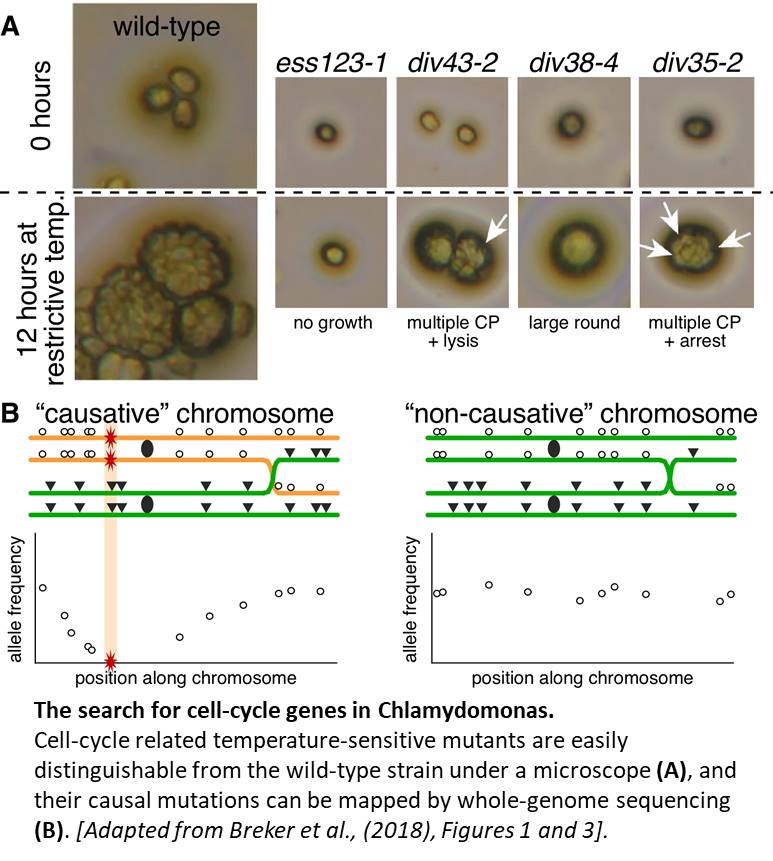
At restrictive temperatures, normal cells grow to many times their initial volume before undergoing several rounds of division, and form clusters easily visualized under a light microscope. Mutants of interest are chosen based on semi-automated microscopy: cells that show no growth defect but fail to form cleavage planes (CPs) between daughter cells or complete division (See Figure, panel A).
But how do you clone the genes? This truly is a case when absence of evidence IS evidence of absence. Mapping mutants is based on co-segregation of molecular markers with the causal mutation in large segregating populations. High-throughput sequencing has been adopted to map mutants quickly, at least in diploid species where setting up crosses and collecting thousands of progeny is easy, like in Arabidopsis (James et al., 2013). But what to do for a haploid organism whose mutants of interest will kill the cells that bear them? Make this a feature of the identification of course!
Mutagen-induced mutations act as markers, and are identified by sequencing pools of mutants following the DNA sudoku principle (Erlich et al., 2009). All mutants, each with a set of unique mutations, are then crossed to other mutants, effectively acting as each other’s wild-type parent (unless they are allelic of course) in mass mating fashion. The pooled progenies are subjected to multiple rounds of selection at the restrictive temperature, and then sequenced at very high coverage. Because the causal mutations are essential for cell survival, they are depleted from the pool (haploid genome, remember?), as well as closely linked mutations, resulting in a distinct pattern in allele frequencies along exactly one chromosome for each mutant (see Figure, panel B).
Following this strategy, Breker and colleagues isolated 350 mutants, 260 of which could be tied down to a single (sometimes two) candidates lesions or genetic loci. ~150 genes were identified (41 of which had more than one allele); Many of them are either mapped to previously described cell cycle genes or can be easily explained to affect division, however a remarkable 47 were found in new genes. Most essential genes identified (75%) have orthologues in the higher plant Arabidopsis, suggesting wide conservation within the plant lineage.
The fun part begins now: what is the function of these genes? At what stage during the cell cycle does each mutant fail? DNA content profiles can provide a first clue; many mutants appear unable to replicate their genomes, while others cannot move beyond DNA replication. Time to divide the work and conquer?
REFERENCES
Breker M, Lieberman K, Cross FR (2018). Comprehensive discovery of cell-cycle-essential pathways in Chlamydomonas reinhardtii. Published May 2018. DOI: https://doi.org/10.1105/tpc.18.00071
Erlich Y, Chang K, Gordon A, Ronen R, Navon O, Rooks M, Hannon GJ (2009). DNA Sudoku–harnessing high-throughput sequencing for multiplexed specimen analysis. Genome Res. 2009 19: 1243-1253.
James GV, Patel V, Nordström KJ, Klasen JR, Salomé PA, Weigel D, Schneeberger K. (2013). User guide for mapping-by-sequencing in Arabidopsis. Genome Biol. 14: R61.
Nowack MK, Harashima H, Dissmeyer N, Zhao X, Bouyer D, Weimer AK, De Winter F, Yang F, Schnittger A. (2012). Genetic framework of cyclin-dependent kinase function in Arabidopsis. Dev Cell. 22: 1030-1040. (2014).