Discovering autophagy protein cargo by protein turnover analysis in Arabidopsis
Li et al. explore the nature of proteins that accumulate in mutants deficient in autophagy machinery.
Plant Cell
Lei Li1,2 and A. Harvey Millar2
- Frontiers Science Center for Cell Responses, Department of Plant Biology and Ecology, College of Life Sciences, Nankai University, 300071 Tianjin, China.
- ARC Centre of Excellence in Plant Energy Biology, School of Molecular Science, The University of Western Australia, 6009 Crawley, WA, Australia.
Background: Autophagy helps maintain the health of cells by degrading cellular components including both functional and structural proteins that are not wanted. If autophagy is inhibited in a plant cell then accumulation of autophagy cargo might be expected. However, proteins can accumulate for a variety of reasons as plants respond to the loss of autophagy; they could be cargo or they could be stress responses or they could be components of alternative pathways for protein degradation. To find which proteins are autophagy cargo, a combined analysis is needed to determine whether a protein is a direct autophagy cargo or an indirect cellular response to autophagy disruption.
Question: Are proteins that accumulate in deficient autophagy mutant genuine autophagy cargo? We wanted to find out a better way to define autophagy protein cargo that did not just measure their abundance and/or presume that transcript abundance indicates protein synthesis rate.
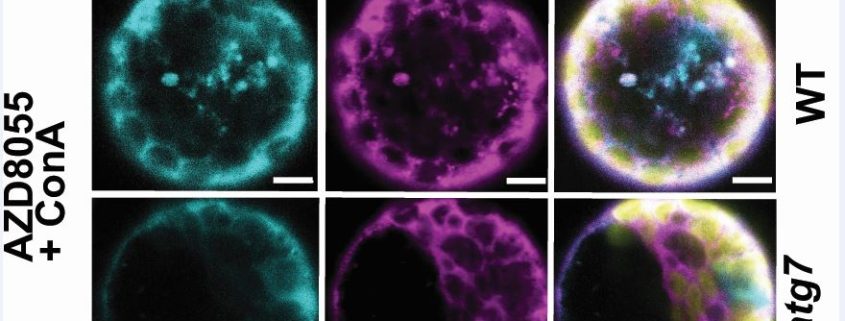
Findings: We find over a hundred proteins that satisfy a new definition of autophagy cargo by both turning over more slowly and accumulating in the Arabidopsis autophagy mutants atg5 and atg11. About half of these proteins are unexpected autophagy cargos including glycolytic enzymes and many other cytosolic proteins. To provide confidence that our way to find new autophagy protein cargos really works, we chose the glycolytic enzyme fructose bisphosphate aldolase 8 (FBA8) to assess by traditional autophagy protein cargo assays. Protoplast assays showed FBA8 tagged with fluorescence can be engulfed by autophagic bodies and delivered to vacuoles for degradation in wildtype but not autophagy mutants. Stopping autophagy by chemical inhibition also stopped FBA8 degradation, supporting our method to find new autophagy targets.
Next steps: While we find a range of new autophagy protein targets, the next step will be to establish how they are recognized by autophagy for delivery to vacuole for proteolysis.
Lei Li, Chun Pong Lee, Xinxin Ding, Yu Qin, Akila Wijerathna-Yapa, Martyna Broda, Marisa S. Otegui, A. Harvey Millar. (2022). Defects in autophagy lead to selective in vivo changes in turnover of cytosolic and organelle proteins in Arabidopsis. https://doi.org/10.1093/plcell/koac185