Commentary: Widespread contamination of Arabidopsis embryo and endosperm transcriptome datasets
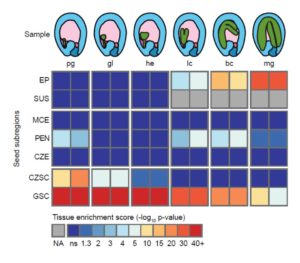 Knowing where a gene is expressed provides valuable information about its function, but that information is compromised if the RNA source is contaminated by other tissues. Schon and Nodine investigated the extent to which Arabidopsis embryo and endosperm transcriptome datasets are affected by tissue contamination. Using seed coat-specific RNAs as markers of maternal tissues and also parent-of-origin small nucleotide polymorphisms (SNPs), the authors found evidence of widespread contamination by maternal tissues, particularly for those datasets representing earlier (smaller) embryos. This finding has implications for interpretations of imprinting (parent-of-origin specific gene expression) in embryos. The authors propose best practices for tissue dissection and also have developed a tool to quantify the extent of contamination of a sample. Plant Cell 10.1105/tpc.16.00845
Knowing where a gene is expressed provides valuable information about its function, but that information is compromised if the RNA source is contaminated by other tissues. Schon and Nodine investigated the extent to which Arabidopsis embryo and endosperm transcriptome datasets are affected by tissue contamination. Using seed coat-specific RNAs as markers of maternal tissues and also parent-of-origin small nucleotide polymorphisms (SNPs), the authors found evidence of widespread contamination by maternal tissues, particularly for those datasets representing earlier (smaller) embryos. This finding has implications for interpretations of imprinting (parent-of-origin specific gene expression) in embryos. The authors propose best practices for tissue dissection and also have developed a tool to quantify the extent of contamination of a sample. Plant Cell 10.1105/tpc.16.00845






Leave a Reply
Want to join the discussion?Feel free to contribute!